A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response
- PMID: 22546241
- PMCID: PMC3446306
- DOI: 10.1186/gb-2012-13-4-r32
A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response
Abstract
Background: Gut microbiota and the host exist in a mutualistic relationship, with the functional composition of the microbiota strongly affecting the health and well-being of the host. Thus, it is important to develop a synthetic approach to study the host transcriptome and the microbiome simultaneously. Early microbial colonization in infants is critically important for directing neonatal intestinal and immune development, and is especially attractive for studying the development of human-commensal interactions. Here we report the results from a simultaneous study of the gut microbiome and host epithelial transcriptome of three-month-old exclusively breast- and formula-fed infants.
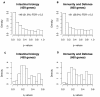
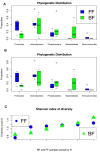
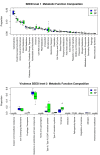
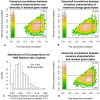
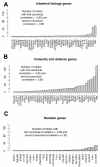
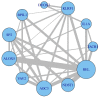
Results: Variation in both host mRNA expression and the microbiome phylogenetic and functional profiles was observed between breast- and formula-fed infants. To examine the interdependent relationship between host epithelial cell gene expression and bacterial metagenomic-based profiles, the host transcriptome and functionally profiled microbiome data were subjected to novel multivariate statistical analyses. Gut microbiota metagenome virulence characteristics concurrently varied with immunity-related gene expression in epithelial cells between the formula-fed and the breast-fed infants.
Conclusions: Our data provide insight into the integrated responses of the host transcriptome and microbiome to dietary substrates in the early neonatal period. We demonstrate that differences in diet can affect, via gut colonization, host expression of genes associated with the innate immune system. Furthermore, the methodology presented in this study can be adapted to assess other host-commensal and host-pathogen interactions using genomic and transcriptomic data, providing a synthetic genomics-based picture of host-commensal relationships.
Figures
Comment in
-
We are what we eat: how the diet of infants affects their gut microbiome.Genome Biol. 2012;13(4):152. doi: 10.1186/gb-2012-13-4-152. Genome Biol. 2012. PMID: 22546514 Free PMC article.
References
-
- Chowdhury SR, King DE, Willing BP, Band MR, Beever JE, Lane AB, Loor JJ, Marini JC, Rund LA, Schook LB, Van Kessel AG, Gaskins HR. Transcriptome profiling of the small intestinal epithelium in germfree versus conventional piglets. BMC Genomics. 2007;8:215. doi: 10.1186/1471-2164-8-215. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

