Cancer detection and biopsy classification using concurrent histopathological and metabolomic analysis of core biopsies
- PMID: 22546470
- PMCID: PMC3446261
- DOI: 10.1186/gm332
Cancer detection and biopsy classification using concurrent histopathological and metabolomic analysis of core biopsies
Abstract
Background: Metabolomics, the non-targeted interrogation of small molecules in a biological sample, is an ideal technology for identifying diagnostic biomarkers. Current tissue extraction protocols involve sample destruction, precluding additional uses of the tissue. This is particularly problematic for high value samples with limited availability, such as clinical tumor biopsies that require structural preservation to histologically diagnose and gauge cancer aggressiveness. To overcome this limitation and increase the amount of information obtained from patient biopsies, we developed and characterized a workflow to perform metabolomic analysis and histological evaluation on the same biopsy sample.
Methods: Biopsies of ten human tissues (muscle, adrenal gland, colon, lung, pancreas, small intestine, spleen, stomach, prostate, kidney) were placed directly in a methanol solution to recover metabolites, precipitate proteins, and fix tissue. Following incubation, biopsies were removed from the solution and processed for histology. Kidney and prostate cancer tumor and benign biopsies were stained with hemotoxylin and eosin and prostate biopsies were subjected to PIN-4 immunohistochemistry. The methanolic extracts were analyzed for metabolites on GC/MS and LC/MS platforms. Raw mass spectrometry data files were automatically extracted using an informatics system that includes peak identification and metabolite identification software.
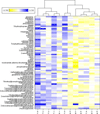
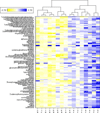
Results: Metabolites across all major biochemical classes (amino acids, peptides, carbohydrates, lipids, nucleotides, cofactors, xenobiotics) were measured. The number (ranging from 260 in prostate to 340 in colon) and identity of metabolites were comparable to results obtained with the current method requiring 30 mg ground tissue. Comparing relative levels of metabolites, cancer tumor from benign kidney and prostate biopsies could be distinguished. Successful histopathological analysis of biopsies by chemical staining (hematoxylin, eosin) and antibody binding (PIN-4, in prostate) showed cellular architecture and immunoreactivity were retained.
Conclusions: Concurrent metabolite extraction and histological analysis of intact biopsies is amenable to the clinical workflow. Methanol fixation effectively preserves a wide range of tissues and is compatible with chemical staining and immunohistochemistry. The method offers an opportunity to augment histopathological diagnosis and tumor classification with quantitative measures of biochemicals in the same tissue sample. Since certain biochemicals have been shown to correlate with disease aggressiveness, this method should prove valuable as an adjunct to differentiate cancer aggressiveness.
Figures






References
-
- Ficarra V, Martignoni G, Maffei N, Brunelli M, Novara G, Zanolla L, Pea M, Artibani W. Original and reviewed nuclear grading according to the Fuhrman system: a multivariate analysis of 388 patients with conventional renal cell carcinoma. Cancer. 2005;103:68–75. doi: 10.1002/cncr.20749. - DOI - PubMed
-
- May M, Brookman-Amissah S, Roigas J, Hartmann A, Storkel S, Kristiansen G, Gilfrich C, Borchardt R, Hoschke B, Kaufmann O, Gunia S. Prognostic accuracy of individual uropathologists in noninvasive urinary bladder carcinoma: a multicentre study comparing the 1973 and 2004 World Health Organisation classifications. Eur Urol. 2010;57:850–858. doi: 10.1016/j.eururo.2009.03.052. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

