Altered Ig hypermutation pattern and frequency in complementary mouse models of DNA polymerase ζ activity
- PMID: 22547703
- PMCID: PMC3358437
- DOI: 10.4049/jimmunol.1102629
Altered Ig hypermutation pattern and frequency in complementary mouse models of DNA polymerase ζ activity
Abstract
To test the hypothesis that DNA polymerase ζ participates in Ig hypermutation, we generated two mouse models of Pol ζ function: a B cell-specific conditional knockout and a knock-in strain with a Pol ζ mutagenesis-enhancing mutation. Pol ζ-deficient B cells had a reduction in mutation frequency at Ig loci in the spleen and in Peyer's patches, whereas knock-in mice with a mutagenic Pol ζ displayed a marked increase in mutation frequency in Peyer's patches, revealing a pattern that was similar to mutations in yeast strains with a homologous mutation in the gene encoding the catalytic subunit of Pol ζ. Combined, these data are best explained by a direct role for DNA polymerase ζ in Ig hypermutation.
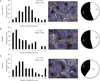
Figures





References
-
- Weigert MG, Cesari IM, Yonkovich SJ, Cohn M. Variability in the lambda light chain sequences of mouse antibody. Nature. 1970;228:1045–1047. - PubMed
-
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. - PubMed
-
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. - PubMed
-
- Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

