One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis
- PMID: 22547828
- PMCID: PMC3384215
- DOI: 10.1073/pnas.1206069109
One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis
Abstract
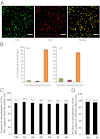
Influenza A virus possesses a segmented genome of eight negative-sense, single-stranded RNAs. The eight segments have been shown to be represented in approximately equal molar ratios in a virus population; however, the exact copy number of each viral RNA segment per individual virus particles has not been determined. We have established an experimental approach based on multicolor single-molecule fluorescent in situ hybridization (FISH) to study the composition of viral RNAs at single-virus particle resolution. Colocalization analysis showed that a high percentage of virus particles package all eight different segments of viral RNAs. To determine the copy number of each RNA segment within individual virus particles, we measured the photobleaching steps of individual virus particles hybridized with fluorescent probes targeting a specific viral RNA. By comparing the photobleaching profiles of probes against the HA RNA segment for the wild-type influenza A/Puerto Rico/8/34 (PR8) and a recombinant PR8 virus carrying two copies of the HA segment, we concluded that only one copy of HA segment is packaged into a wild type virus particle. Our results showed similar photobleaching behaviors for other RNA segments, suggesting that for the majority of the virus particles, only one copy of each RNA segment is packaged into one virus particle. Together, our results support that the packaging of influenza viral genome is a selective process.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Packaging of influenza virus genome: robustness of selection.Proc Natl Acad Sci U S A. 2012 Jun 5;109(23):8797-8. doi: 10.1073/pnas.1206736109. Epub 2012 May 21. Proc Natl Acad Sci U S A. 2012. PMID: 22615368 Free PMC article. No abstract available.
References
-
- Palese P, Shaw ML. Orthomyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Field’s Virology. Vol 2. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1648–1689.
-
- Enami M, Sharma G, Benham C, Palese P. An influenza virus containing nine different RNA segments. Virology. 1991;185:291–298. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

