Resistance of Subtype C HIV-1 Strains to Anti-V3 Loop Antibodies
- PMID: 22548061
- PMCID: PMC3323838
- DOI: 10.1155/2012/803535
Resistance of Subtype C HIV-1 Strains to Anti-V3 Loop Antibodies
Abstract
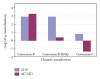
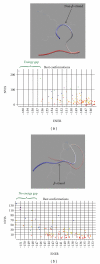
HIV-1's subtype C V3 loop consensus sequence exhibits increased resistance to anti-V3 antibody-mediated neutralization as compared to the subtype B consensus sequence. The dynamic 3D structure of the consensus C V3 loop crown, visualized by ab initio folding, suggested that the resistance derives from structural rigidity and non-β-strand secondary protein structure in the N-terminal strand of the β-hairpin of the V3 loop crown, which is where most known anti-V3 loop antibodies bind. The observation of either rigidity or non-β-strand structure in this region correlated with observed resistance to antibody-mediated neutralization in a series of chimeric pseudovirus (psV) mutants. The results suggest the presence of an epitope-independent, neutralization-relevant structural difference in the antibody-targeted region of the V3 loop crown between subtype C and subtype B, a difference that we hypothesize may contribute to the divergent pattern of global spread between these subtypes. As antibodies to a variable loop were recently identified as an inverse correlate of risk for HIV infection, the structure-function relationships discussed in this study may have relevance to HIV vaccine research.
Figures
References
-
- Goudsmit J, Debouck C, Meloen RH, et al. Human immunodeficiency virus type 1 neutralization epitope with conserved architecture elicits early type-specific antibodies in experimentally infected chimpanzees. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(12):4478–4482. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

