Identification of unique MEK-dependent genes in GNAQ mutant uveal melanoma involved in cell growth, tumor cell invasion, and MEK resistance
- PMID: 22550165
- PMCID: PMC3433236
- DOI: 10.1158/1078-0432.CCR-11-3086
Identification of unique MEK-dependent genes in GNAQ mutant uveal melanoma involved in cell growth, tumor cell invasion, and MEK resistance
Abstract
Purpose: Metastatic uveal melanoma represents the most common intraocular malignancy with very poor prognosis and no effective treatments. Oncogenic mutations in the G-protein α-subunit q and 11 have been described in about 85% of uveal melanomas and confer constitutive activation. Multiple signaling pathways are induced as a consequence of GNAQ/11 activation, which include the MEK/ERK kinase cascade. We analyzed the transcriptional profile of cell lines treated with a mitogen-activated protein (MAP)/extracellular signal-regulated (ERK) kinase (MEK) inhibitor to identify gene targets of activated GNAQ and to evaluate the biologic importance of these genes in uveal melanoma.
Experimental design: We conducted microarray analysis of uveal melanoma cell lines with GNAQ mutations treated with the MEK inhibitor selumetinib. For comparison, we used cells carrying BRAF(V600E) and cells without either mutation. Changes in the expression of selected genes were then confirmed by quantitative real-time PCR and immunoblotting.
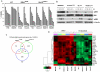
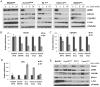
Results: We found that GNAQ mutant cells have a MEK-dependent transcriptional output and identified a unique set of genes that are downregulated by MEK inhibition, including the RNA helicase DDX21 and the cyclin-dependent kinase regulator CDK5R1 whereas Jun was induced. We provide evidence that these genes are involved in cell proliferation, tumor cell invasion, and drug resistance, respectively. Furthermore, we show that selumetinib treatment regulates the expression of these genes in tumor tissues of patients with metastatic GNAQ/11 mutant uveal melanoma.
Conclusions: Our findings define a subset of transcriptionally regulated genes by selumetinib in GNAQ mutant cells and provide new insights into understanding the biologic effect of MEK inhibition in this disease.
©2012 AACR.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

