LDGIdb: a database of gene interactions inferred from long-range strong linkage disequilibrium between pairs of SNPs
- PMID: 22551073
- PMCID: PMC3441865
- DOI: 10.1186/1756-0500-5-212
LDGIdb: a database of gene interactions inferred from long-range strong linkage disequilibrium between pairs of SNPs
Abstract
Background: Complex human diseases may be associated with many gene interactions. Gene interactions take several different forms and it is difficult to identify all of the interactions that are potentially associated with human diseases. One approach that may fill this knowledge gap is to infer previously unknown gene interactions via identification of non-physical linkages between different mutations (or single nucleotide polymorphisms, SNPs) to avoid hitchhiking effect or lack of recombination. Strong non-physical SNP linkages are considered to be an indication of biological (gene) interactions. These interactions can be physical protein interactions, regulatory interactions, functional compensation/antagonization or many other forms of interactions. Previous studies have shown that mutations in different genes can be linked to the same disorders. Therefore, non-physical SNP linkages, coupled with knowledge of SNP-disease associations may shed more light on the role of gene interactions in human disorders. A user-friendly web resource that integrates information about non-physical SNP linkages, gene annotations, SNP information, and SNP-disease associations may thus be a good reference for biomedical research.
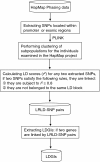
Findings: Here we extracted the SNPs located within the promoter or exonic regions of protein-coding genes from the HapMap database to construct a database named the Linkage-Disequilibrium-based Gene Interaction database (LDGIdb). The database stores 646,203 potential human gene interactions, which are potential interactions inferred from SNP pairs that are subject to long-range strong linkage disequilibrium (LD), or non-physical linkages. To minimize the possibility of hitchhiking, SNP pairs inferred to be non-physically linked were required to be located in different chromosomes or in different LD blocks of the same chromosomes. According to the genomic locations of the involved SNPs (i.e., promoter, untranslated region (UTR) and coding region (CDS)), the SNP linkages inferred were categorized into promoter-promoter, promoter-UTR, promoter-CDS, CDS-CDS, CDS-UTR and UTR-UTR linkages. For the CDS-related linkages, the coding SNPs were further classified into nonsynonymous and synonymous variations, which represent potential gene interactions at the protein and RNA level, respectively. The LDGIdb also incorporates human disease-association databases such as Genome-Wide Association Studies (GWAS) and Online Mendelian Inheritance in Man (OMIM), so that the user can search for potential disease-associated SNP linkages. The inferred SNP linkages are also classified in the context of population stratification to provide a resource for investigating potential population-specific gene interactions.
Conclusion: The LDGIdb is a user-friendly resource that integrates non-physical SNP linkages and SNP-disease associations for studies of gene interactions in human diseases. With the help of the LDGIdb, it is plausible to infer population-specific SNP linkages for more focused studies, an avenue that is potentially important for pharmacogenetics. Moreover, by referring to disease-association information such as the GWAS data, the LDGIdb may help identify previously uncharacterized disease-associated gene interactions and potentially lead to new discoveries in studies of human diseases.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

