Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments
- PMID: 22551871
- PMCID: PMC3449235
- DOI: 10.1038/ismej.2012.37
Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments
Abstract
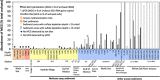
Members of the highly diverse Miscellaneous Crenarchaeotal Group (MCG) are globally distributed in various marine and continental habitats. In this study, we applied a polyphasic approach (rRNA slot blot hybridization, quantitative PCR (qPCR) and catalyzed reporter deposition FISH) using newly developed probes and primers for the in situ detection and quantification of MCG crenarchaeota in diverse types of marine sediments and microbial mats. In general, abundance of MCG (cocci, 0.4 μm) relative to other archaea was highest (12-100%) in anoxic, low-energy environments characterized by deeper sulfate depletion and lower microbial respiration rates (P=0.06 for slot blot and P=0.05 for qPCR). When studied in high depth resolution in the White Oak River estuary and Hydrate Ridge methane seeps, changes in MCG abundance relative to total archaea and MCG phylogenetic composition did not correlate with changes in sulfate reduction or methane oxidation with depth. In addition, MCG abundance did not vary significantly (P>0.1) between seep sites (with high rates of methanotrophy) and non-seep sites (with low rates of methanotrophy). This suggests that MCG are likely not methanotrophs. MCG crenarchaeota are highly diverse and contain 17 subgroups, with a range of intragroup similarity of 82 to 94%. This high diversity and widespread distribution in subsurface sediments indicates that this group is globally important in sedimentary processes.
Figures





References
-
- Boetius A, Ravenschlag K, Schubert C, Rickert D, Widdel F, Gieseke A, et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. - PubMed
-
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

