Histology-specific microRNA alterations in melanoma
- PMID: 22551973
- PMCID: PMC3648670
- DOI: 10.1038/jid.2011.451
Histology-specific microRNA alterations in melanoma
Abstract
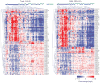
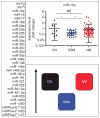
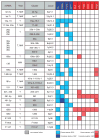
We examined the microRNA signature that distinguishes the most common melanoma histological subtypes, superficial spreading melanoma (SSM) and nodular melanoma (NM). We also investigated the mechanisms underlying the differential expression of histology-specific microRNAs. MicroRNA array performed on a training cohort of 82 primary melanoma tumors (26 SSM, 56 NM), and nine congenital nevi (CN) revealed 134 microRNAs differentially expressed between SSM and NM (P<0.05). Out of 134 microRNAs, 126 remained significant after controlling for thickness and 31 were expressed at a lower level in SSM compared with both NM and CN. For seven microRNAs (let-7g, miR-15a, miR-16, miR-138, miR-181a, miR-191, and miR-933), the downregulation was associated with selective genomic loss in SSM cell lines and primary tumors, but not in NM cell lines and primary tumors. The lower expression level of six out of seven microRNAs in SSM compared with NM was confirmed by real-time PCR on a subset of cases in the training cohort and validated in an independent cohort of 97 melanoma cases (38 SSM, 59 NM). Our data support a molecular classification in which SSM and NM are two molecularly distinct phenotypes. Therapeutic strategies that take into account subtype-specific alterations might improve the outcome of melanoma patients.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Baumert J, Schmidt M, Giehl KA, et al. Time trends in tumour thickness vary in subgroups: analysis of 6475 patients by age, tumour site and melanoma subtype. Melanoma Res. 2009;19:24–30. - PubMed
-
- Caramuta S, Egyhazi S, Rodolfo M, et al. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130:2062–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

