EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes
- PMID: 22552007
- PMCID: PMC3437410
- DOI: 10.1038/leu.2012.120
EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes
Abstract
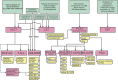
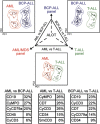
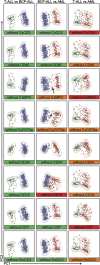
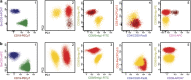
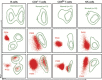
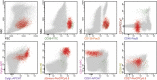
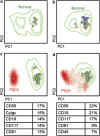
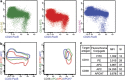
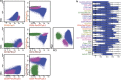
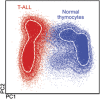
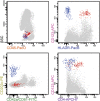
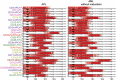
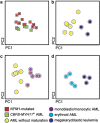
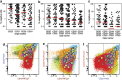
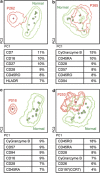
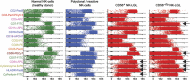
Most consensus leukemia & lymphoma antibody panels consist of lists of markers based on expert opinions, but they have not been validated. Here we present the validated EuroFlow 8-color antibody panels for immunophenotyping of hematological malignancies. The single-tube screening panels and multi-tube classification panels fit into the EuroFlow diagnostic algorithm with entries defined by clinical and laboratory parameters. The panels were constructed in 2-7 sequential design-evaluation-redesign rounds, using novel Infinicyt software tools for multivariate data analysis. Two groups of markers are combined in each 8-color tube: (i) backbone markers to identify distinct cell populations in a sample, and (ii) markers for characterization of specific cell populations. In multi-tube panels, the backbone markers were optimally placed at the same fluorochrome position in every tube, to provide identical multidimensional localization of the target cell population(s). The characterization markers were positioned according to the diagnostic utility of the combined markers. Each proposed antibody combination was tested against reference databases of normal and malignant cells from healthy subjects and WHO-based disease entities, respectively. The EuroFlow studies resulted in validated and flexible 8-color antibody panels for multidimensional identification and characterization of normal and aberrant cells, optimally suited for immunophenotypic screening and classification of hematological malignancies.
Figures














References
-
- Foon KA, Todd RF. Immunologic classification of leukemia and lymphoma. Blood. 1986;68:1–31. - PubMed
-
- van Dongen JJ, Adriaansen HJ, Hooijkaas H. Immunophenotyping of leukaemias and non-Hodgkin's lymphomas. Immunological markers and their CD codes. Neth J Med. 1988;33:298–314. - PubMed
-
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues4th edn.International Agency for Research on Cancer: Lyon; 2008. pp 439.
-
- Macedo A, Orfao A, Vidriales MB, Lopez-Berges MC, Valverde B, Gonzalez M, et al. Characterization of aberrant phenotypes in acute myeloblastic leukemia. Ann Hematol. 1995;70:189–194. - PubMed
-
- Lucio P, Parreira A, van den Beemd MW, van Lochem EG, van Wering ER, Baars E, et al. Flow cytometric analysis of normal B cell differentiation: a frame of reference for the detection of minimal residual disease in precursor-B-ALL. Leukemia. 1999;13:419–427. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

