Accumulation of multipotent progenitors with a basal differentiation bias during aging of human mammary epithelia
- PMID: 22552289
- PMCID: PMC3399034
- DOI: 10.1158/0008-5472.CAN-12-0157
Accumulation of multipotent progenitors with a basal differentiation bias during aging of human mammary epithelia
Abstract
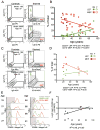
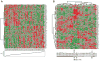
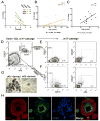
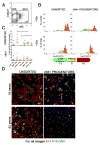
Women older than 50 years account for 75% of new breast cancer diagnoses, and the majority of these tumors are of a luminal subtype. Although age-associated changes, including endocrine profiles and alterations within the breast microenvironment, increase cancer risk, an understanding of the cellular and molecular mechanisms that underlies these observations is lacking. In this study, we generated a large collection of normal human mammary epithelial cell strains from women ages 16 to 91 years, derived from primary tissues, to investigate the molecular changes that occur in aging breast cells. We found that in finite lifespan cultured and uncultured epithelial cells, aging is associated with a reduction of myoepithelial cells and an increase in luminal cells that express keratin 14 and integrin-α6, a phenotype that is usually expressed exclusively in myoepithelial cells in women younger than 30 years. Changes to the luminal lineage resulted from age-dependent expansion of defective multipotent progenitors that gave rise to incompletely differentiated luminal or myoepithelial cells. The aging process therefore results in both a shift in the balance of luminal/myoepithelial lineages and to changes in the functional spectrum of multipotent progenitors, which together increase the potential for malignant transformation. Together, our findings provide a cellular basis to explain the observed vulnerability to breast cancer that increases with age.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- DePinho RA. The age of cancer. Nature. 2000;408(6809):248–54. - PubMed
-
- Jemal A, et al. Cancer statistics, 2007. CA Cancer J Clin. 2007;57(1):43–66. - PubMed
-
- Smigal C, et al. Trends in breast cancer by race and ethnicity: update 2006. CA Cancer J Clin. 2006;56(3):168–83. - PubMed
-
- Carey LA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–502. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

