Total centromere size and genome size are strongly correlated in ten grass species
- PMID: 22552915
- PMCID: PMC3391362
- DOI: 10.1007/s10577-012-9284-1
Total centromere size and genome size are strongly correlated in ten grass species
Abstract
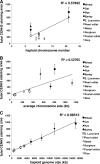
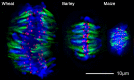
It has been known for decades that centromere size varies across species, but the factors involved in setting centromere boundaries are unknown. As a means to address this question, we estimated centromere sizes in ten species of the grass family including rice, maize, and wheat, which diverged 60~80 million years ago and vary by 40-fold in genome size. Measurements were made using a broadly reactive antibody to rice centromeric histone H3 (CENH3). In species-wide comparisons, we found a clear linear relationship between total centromere size and genome size. Species with large genomes and few chromosomes tend to have the largest centromeres (e.g., rye) while species with small genomes and many chromosomes have the smallest centromeres (e.g., rice). However, within a species, centromere size is surprisingly uniform. We present evidence from three oat-maize addition lines that support this claim, indicating that each of three maize centromeres propagated in oat are not measurably different from each other. In the context of previously published data, our results suggest that the apparent correlation between chromosome and centromere size is incidental to a larger trend that reflects genome size. Centromere size may be determined by a limiting component mechanism similar to that described for Caenorhabditis elegans centrosomes.
Figures



References
-
- Bennetzen JL, Freeling M. The unified grass genome: synergy in synteny. Genome Res. 1997;7:301–306. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

