Methods for simultaneous EEG-fMRI: an introductory review
- PMID: 22553012
- PMCID: PMC6622140
- DOI: 10.1523/JNEUROSCI.0447-12.2012
Methods for simultaneous EEG-fMRI: an introductory review
Abstract
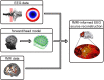
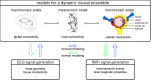
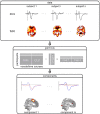
The simultaneous recording and analysis of electroencephalography (EEG) and fMRI data in human systems, cognitive and clinical neurosciences is rapidly evolving and has received substantial attention. The significance of multimodal brain imaging is documented by a steadily increasing number of laboratories now using simultaneous EEG-fMRI aiming to achieve both high temporal and spatial resolution of human brain function. Due to recent developments in technical and algorithmic instrumentation, the rate-limiting step in multimodal studies has shifted from data acquisition to analytic aspects. Here, we introduce and compare different methods for data integration and identify the benefits that come with each approach, guiding the reader toward an understanding and informed selection of the integration approach most suitable for addressing a particular research question.
Figures

References
-
- Babiloni F, Carducci F, Cincotti F, Del Gratta C, Roberti GM, Romani GL, Rossini PM, Babiloni C. Integration of high resolution EEG and functional magnetic resonance in the study of human movement-related potentials. Methods Inf Med. 2000;39:179–182. - PubMed
-
- Babiloni F, Babiloni C, Carducci F, Del Gratta C, Romani GL, Rossini PM, Cincotti F. Cortical source estimate of combined high resolution EEG and fMRI data related to voluntary movements. Methods Inf Med. 2002;41:443–450. - PubMed
-
- Bell AJ, Sejnowski TJ. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 1995;7:1129–1159. - PubMed
-
- Bénar CG, Schön D, Grimault S, Nazarian B, Burle B, Roth M, Badier JM, Marquis P, Liegeois-Chauvel C, Anton JL. Single-trial analysis of oddball event-related potentials in simultaneous EEG-fMRI. Hum Brain Mapp. 2007;28:602–613. - PubMed
-
- Besserve M, Jerbi K, Laurent F, Baillet S, Martinerie J, Garnero L. Classification methods for ongoing EEG and MEG signals. Biol Res. 2007;40:415–437. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
