Tbr2 is essential for hippocampal lineage progression from neural stem cells to intermediate progenitors and neurons
- PMID: 22553033
- PMCID: PMC3366485
- DOI: 10.1523/JNEUROSCI.0532-12.2012
Tbr2 is essential for hippocampal lineage progression from neural stem cells to intermediate progenitors and neurons
Abstract
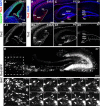
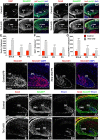
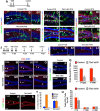
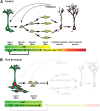
Neurogenesis in the dentate gyrus has been implicated in cognitive functions, including learning and memory, and may be abnormal in major neuropsychiatric disorders, such as depression. Dentate neurogenesis is regulated by interactions between extrinsic factors and intrinsic transcriptional cascades that are currently not well understood. Here we show that Tbr2 (also known as Eomes), a T-box transcription factor expressed by intermediate neuronal progenitors (INPs), is critically required for neurogenesis in the dentate gyrus of developing and adult mice. In the absence of Tbr2, INPs are depleted despite augmented neural stem cell (NSC) proliferation, and neurogenesis is halted as the result of failed neuronal differentiation. Interestingly, we find that Tbr2 likely promotes lineage progression from NSC to neuronal-specified INP in part by repression of Sox2, a key determinant of NSC identity. These findings suggest that Tbr2 expression in INPs is critical for neuronal differentiation in the dentate gyrus and that INPs are an essential stage in the lineage from NSCs to new granule neurons in the dentate gyrus.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
