From sequence to molecular pathology, and a mechanism driving the neuroendocrine phenotype in prostate cancer
- PMID: 22553170
- PMCID: PMC3659819
- DOI: 10.1002/path.4047
From sequence to molecular pathology, and a mechanism driving the neuroendocrine phenotype in prostate cancer
Abstract
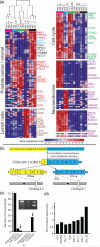
The current paradigm of cancer care relies on predictive nomograms which integrate detailed histopathology with clinical data. However, when predictions fail, the consequences for patients are often catastrophic, especially in prostate cancer where nomograms influence the decision to therapeutically intervene. We hypothesized that the high dimensional data afforded by massively parallel sequencing (MPS) is not only capable of providing biological insights, but may aid molecular pathology of prostate tumours. We assembled a cohort of six patients with high-risk disease, and performed deep RNA and shallow DNA sequencing in primary tumours and matched metastases where available. Our analysis identified copy number abnormalities, accurately profiled gene expression levels, and detected both differential splicing and expressed fusion genes. We revealed occult and potentially dormant metastases, unambiguously supporting the patients' clinical history, and implicated the REST transcriptional complex in the development of neuroendocrine prostate cancer, validating this finding in a large independent cohort. We massively expand on the number of novel fusion genes described in prostate cancer; provide fresh evidence for the growing link between fusion gene aetiology and gene expression profiles; and show the utility of fusion genes for molecular pathology. Finally, we identified chromothripsis in a patient with chronic prostatitis. Our results provide a strong foundation for further development of MPS-based molecular pathology.
Copyright © 2012 Pathological Society of Great Britain and Ireland. Published by John Wiley & Sons, Ltd.
Figures




References
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics. Ca Cancer J Clin 2012. 2012;62:10–29. - PubMed
-
- Graefen M, Karakiewicz PI, Cagiannos I, et al. Validation study of the accuracy of a postoperative nomogram for recurrence after radical prostatectomy for localized prostate cancer. J Clin Oncol. 2002;20:951–956. - PubMed
-
- Graefen M, Karakiewicz PI, Cagiannos I, et al. International validation of a preoperative nomogram for prostate cancer recurrence after radical prostatectomy. J Clin Oncol. 2002;20:3206–3212. - PubMed
-
- Kattan MW, Eastham JA, Stapleton AM, et al. A preoperative nomogram for disease recurrence following radical prostatectomy for prostate cancer. J Natl Cancer Inst. 1998;90:766–771. - PubMed
-
- Kattan MW, Wheeler TM, Scardino PT. Postoperative nomogram for disease recurrence after radical prostatectomy for prostate cancer. J Clin Oncol. 1999;17:1499–1507. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

