Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus
- PMID: 22553322
- PMCID: PMC3416313
- DOI: 10.1128/JVI.00157-12
Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus
Abstract
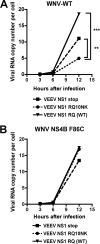
Flavivirus NS1 is a nonstructural glycoprotein that is expressed on the cell surface and secreted into the extracellular space. Despite its transit through the secretory pathway, NS1 is an essential gene linked to early viral RNA replication. How this occurs has remained a mystery given the disparate localization of NS1 and the viral RNA replication complex, as the latter is present on the cytosolic face of the endoplasmic reticulum (ER). We recently identified an N-terminal di-amino acid motif in NS1 that modulates protein targeting and affected viral replication. Exchange of two amino acids at positions 10 and 11 from dengue virus (DENV) into West Nile virus (WNV) NS1 (RQ10NK) changed its relative surface expression and secretion and attenuated infectivity. However, the phenotype of WNV containing NS1 RQ10NK was unstable, as within two passages heterogeneous plaque variants were observed. Here, using a mutant WNV encoding the NS1 RQ10NK mutation, we identified a suppressor mutation (F86C) in NS4B, a virally encoded transmembrane protein with loops on both the luminal and cytoplasmic sides of the ER membrane. Introduction of NS4B F86C specifically rescued RNA replication of mutant WNV but did not affect the wild-type virus. Mass spectrometry and coimmunoprecipitation studies established a novel physical interaction between NS1 and NS4B, suggesting a mechanism for how luminal NS1 conveys signals to the cytoplasm to regulate RNA replication.
Figures



Similar articles
-
A short N-terminal peptide motif on flavivirus nonstructural protein NS1 modulates cellular targeting and immune recognition.J Virol. 2010 Sep;84(18):9516-32. doi: 10.1128/JVI.00775-10. Epub 2010 Jun 30. J Virol. 2010. PMID: 20592095 Free PMC article.
-
Non-structural protein-1 is required for West Nile virus replication complex formation and viral RNA synthesis.Virol J. 2013 Nov 18;10:339. doi: 10.1186/1743-422X-10-339. Virol J. 2013. PMID: 24245822 Free PMC article.
-
Levels of Circulating NS1 Impact West Nile Virus Spread to the Brain.J Virol. 2021 Sep 27;95(20):e0084421. doi: 10.1128/JVI.00844-21. Epub 2021 Aug 4. J Virol. 2021. PMID: 34346770 Free PMC article.
-
Replication cycle and molecular biology of the West Nile virus.Viruses. 2013 Dec 27;6(1):13-53. doi: 10.3390/v6010013. Viruses. 2013. PMID: 24378320 Free PMC article. Review.
-
Structure and function of the parvoviral NS1 protein: a review.Virus Genes. 2023 Apr;59(2):195-203. doi: 10.1007/s11262-022-01944-2. Epub 2022 Oct 17. Virus Genes. 2023. PMID: 36253516 Review.
Cited by
-
Increased early RNA replication by chimeric West Nile virus W956IC leads to IPS-1-mediated activation of NF-κB and insufficient virus-mediated counteraction of the resulting canonical type I interferon signaling.J Virol. 2013 Jul;87(14):7952-65. doi: 10.1128/JVI.02842-12. Epub 2013 May 15. J Virol. 2013. PMID: 23678179 Free PMC article.
-
K48-linked polyubiquitination of dengue virus NS1 protein inhibits its interaction with the viral partner NS4B.Virus Res. 2018 Feb 15;246:1-11. doi: 10.1016/j.virusres.2017.12.013. Epub 2017 Dec 30. Virus Res. 2018. PMID: 29294313 Free PMC article.
-
A Sensitive Yellow Fever Virus Entry Reporter Identifies Valosin-Containing Protein (VCP/p97) as an Essential Host Factor for Flavivirus Uncoating.mBio. 2020 Apr 14;11(2):e00467-20. doi: 10.1128/mBio.00467-20. mBio. 2020. PMID: 32291299 Free PMC article.
-
Dengue Virus Non-structural Protein 1 Modulates Infectious Particle Production via Interaction with the Structural Proteins.PLoS Pathog. 2015 Nov 12;11(11):e1005277. doi: 10.1371/journal.ppat.1005277. eCollection 2015. PLoS Pathog. 2015. PMID: 26562291 Free PMC article.
-
Functional Roles and Host Interactions of Orthoflavivirus Non-Structural Proteins During Replication.Pathogens. 2025 Feb 12;14(2):184. doi: 10.3390/pathogens14020184. Pathogens. 2025. PMID: 40005559 Free PMC article. Review.
References
-
- Avirutnan P, et al. 2007. Secreted NS1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate E. PLoS Pathog. 3:e183 doi:10.1371/journal.ppat.0030183 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

