An in-frame deletion in the NS protein-coding sequence of parvovirus H-1PV efficiently stimulates export and infectivity of progeny virions
- PMID: 22553326
- PMCID: PMC3416301
- DOI: 10.1128/JVI.00212-12
An in-frame deletion in the NS protein-coding sequence of parvovirus H-1PV efficiently stimulates export and infectivity of progeny virions
Abstract
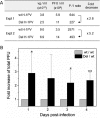
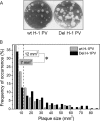
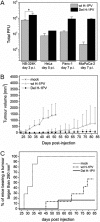
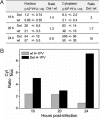
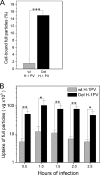
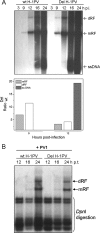
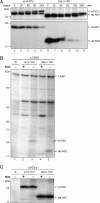
An in-frame, 114-nucleotide-long deletion that affects the NS-coding sequence was created in the infectious molecular clone of the standard parvovirus H-1PV, thereby generating Del H-1PV. The plasmid was transfected and further propagated in permissive human cell lines in order to analyze the effects of the deletion on virus fitness. Our results show key benefits of this deletion, as Del H-1PV proved to exhibit (i) higher infectivity (lower particle-to-infectivity ratio) in vitro and (ii) enhanced tumor growth suppression in vivo compared to wild-type H-1PV. This increased infectivity correlated with an accelerated egress of Del H-1PV progeny virions in producer cells and with an overall stimulation of the viral life cycle in subsequently infected cells. Indeed, virus adsorption and internalization were significantly improved with Del H-1PV, which may account for the earlier appearance of viral DNA replicative forms that was observed with Del H-1PV than wild-type H-1PV. We hypothesize that the internal deletion within the NS2 and/or NS1 protein expressed by Del H-1PV results in the stimulation of some step(s) of the viral life cycle, in particular, a maturation step(s), leading to more efficient nuclear export of infectious viral particles and increased fitness of the virus produced.
Figures

Similar articles
-
Mutations in the Non-Structural Protein-Coding Sequence of Protoparvovirus H-1PV Enhance the Fitness of the Virus and Show Key Benefits Regarding the Transduction Efficiency of Derived Vectors.Viruses. 2018 Mar 27;10(4):150. doi: 10.3390/v10040150. Viruses. 2018. PMID: 29584637 Free PMC article.
-
Through its nonstructural protein NS1, parvovirus H-1 induces apoptosis via accumulation of reactive oxygen species.J Virol. 2010 Jun;84(12):5909-22. doi: 10.1128/JVI.01797-09. Epub 2010 Apr 7. J Virol. 2010. PMID: 20375165 Free PMC article.
-
SMAD4: a predictive marker of PDAC cell permissiveness for oncolytic infection with parvovirus H-1PV.Int J Cancer. 2010 Jun 15;126(12):2914-27. doi: 10.1002/ijc.24992. Int J Cancer. 2010. PMID: 19856310
-
Double-faceted mechanism of parvoviral oncosuppression.Curr Opin Virol. 2015 Aug;13:17-24. doi: 10.1016/j.coviro.2015.03.008. Epub 2015 Apr 2. Curr Opin Virol. 2015. PMID: 25841215 Review.
-
Virotherapy of digestive tumors with rodent parvovirus: overview and perspectives.Expert Opin Biol Ther. 2016;16(5):645-53. doi: 10.1517/14712598.2016.1151492. Epub 2016 Mar 28. Expert Opin Biol Ther. 2016. PMID: 26855087 Review.
Cited by
-
H-1 Parvovirus as a Cancer-Killing Agent: Past, Present, and Future.Viruses. 2019 Jun 18;11(6):562. doi: 10.3390/v11060562. Viruses. 2019. PMID: 31216641 Free PMC article. Review.
-
Genome sequence of tumor virus x, a member of the genus protoparvovirus in the family parvoviridae.Genome Announc. 2014 Jul 31;2(4):e00758-14. doi: 10.1128/genomeA.00758-14. Genome Announc. 2014. PMID: 25081268 Free PMC article.
-
Natural Product Bruceine A from (L.) Merr. as a Potential LDLR Inhibitor That Facilitates Antiviral Effect.ACS Omega. 2025 Jun 24;10(26):28210-28219. doi: 10.1021/acsomega.5c02956. eCollection 2025 Jul 8. ACS Omega. 2025. PMID: 40657109 Free PMC article.
-
Mutations in the Non-Structural Protein-Coding Sequence of Protoparvovirus H-1PV Enhance the Fitness of the Virus and Show Key Benefits Regarding the Transduction Efficiency of Derived Vectors.Viruses. 2018 Mar 27;10(4):150. doi: 10.3390/v10040150. Viruses. 2018. PMID: 29584637 Free PMC article.
-
Preclinical Testing of an Oncolytic Parvovirus: Standard Protoparvovirus H-1PV Efficiently Induces Osteosarcoma Cell Lysis In Vitro.Viruses. 2017 Oct 17;9(10):301. doi: 10.3390/v9100301. Viruses. 2017. PMID: 29039746 Free PMC article.
References
-
- Baer S, Daeffler L, Rommelaere J, Nueesch JPF. 2008. Vesicular egress of non-enveloped lytic parvoviruses depends on gelsolin functioning. PLoS Pathog. 4:e1000126 doi:10.1371/journal.ppat.1000126 - DOI - PMC - PubMed
-
- Besselsen DG, et al. 1996. Molecular characterization of newly recognized rodent parvoviruses. J. Gen. Virol. 77(Pt 5):899–911 - PubMed
-
- Besselsen DG, Romero MJ, Wagner AM, Henderson KS, Livingston RS. 2006. Identification of novel murine parvovirus strains by epidemiological analysis of naturally infected mice. J. Gen. Virol. 87:1543–1556 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

