Adaptor protein 2-mediated endocytosis of the β-secretase BACE1 is dispensable for amyloid precursor protein processing
- PMID: 22553349
- PMCID: PMC3374752
- DOI: 10.1091/mbc.E11-11-0944
Adaptor protein 2-mediated endocytosis of the β-secretase BACE1 is dispensable for amyloid precursor protein processing
Erratum in
- Mol Biol Cell. 2012 Jul;23(13):2620
Abstract
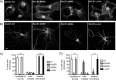
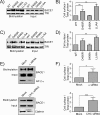
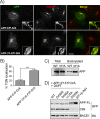
The β-site amyloid precursor protein (APP)-cleaving enzyme 1 (BACE1) is a transmembrane aspartyl protease that catalyzes the proteolytic processing of APP and other plasma membrane protein precursors. BACE1 cycles between the trans-Golgi network (TGN), the plasma membrane, and endosomes by virtue of signals contained within its cytosolic C-terminal domain. One of these signals is the DXXLL-motif sequence DISLL, which controls transport between the TGN and endosomes via interaction with GGA proteins. Here we show that the DISLL sequence is embedded within a longer [DE]XXXL[LI]-motif sequence, DDISLL, which mediates internalization from the plasma membrane by interaction with the clathrin-associated, heterotetrameric adaptor protein 2 (AP-2) complex. Mutation of this signal or knockdown of either AP-2 or clathrin decreases endosomal localization and increases plasma membrane localization of BACE1. Remarkably, internalization-defective BACE1 is able to cleave an APP mutant that itself cannot be delivered to endosomes. The drug brefeldin A reversibly prevents BACE1-catalyzed APP cleavage, ruling out that this reaction occurs in the endoplasmic reticulum (ER) or ER-Golgi intermediate compartment. Taken together, these observations support the notion that BACE1 is capable of cleaving APP in late compartments of the secretory pathway.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

