Evolutionary constraints on the plastid tRNA set decoding methionine and isoleucine
- PMID: 22553362
- PMCID: PMC3413147
- DOI: 10.1093/nar/gks350
Evolutionary constraints on the plastid tRNA set decoding methionine and isoleucine
Abstract
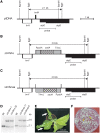
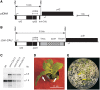
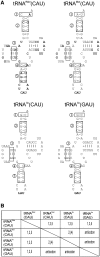
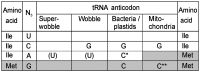
The plastid (chloroplast) genomes of seed plants typically encode 30 tRNAs. Employing wobble and superwobble mechanisms, most codon boxes are read by only one or two tRNA species. The reduced set of plastid tRNAs follows the evolutionary trend of organellar genomes to shrink in size and coding capacity. A notable exception is the AUN codon box specifying methionine and isoleucine, which is decoded by four tRNA species in nearly all seed plants. However, three of these four tRNA genes were lost from the genomes of some parasitic plastid-containing lineages, possibly suggesting that less than four tRNA species could be sufficient to decode the triplets in the AUN box. To test this hypothesis, we have performed knockout experiments for the four AUN-decoding tRNAs in tobacco (Nicotiana tabacum) plastids. We find that all four tRNA genes are essential under both autotrophic and heterotrophic growth conditions, possibly suggesting tRNA import into plastids of parasitic plastid-bearing species. Phylogenetic analysis of the four plastid tRNA genes reveals striking conservation of all those bacterial features that are involved in discrimination between the different tRNA species containing CAU anticodons.
Figures



References
-
- Crick FHC. Codon-anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966;19:548–555. - PubMed
-
- Sugiura M, Wakasugi T. Compilation and comparison of transfer RNA genes from tobacco chloroplasts. Crit. Rev. Plant Sci. 1989;8:89–101.
-
- Bock R. Structure, function, and inheritance of plastid genomes. Top. Curr. Genet. 2007;19:29–63.
-
- Rogalski M, Karcher D, Bock R. Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol. 2008;15:192–198. - PubMed

