CSA: comprehensive comparison of pairwise protein structure alignments
- PMID: 22553365
- PMCID: PMC3394275
- DOI: 10.1093/nar/gks362
CSA: comprehensive comparison of pairwise protein structure alignments
Abstract
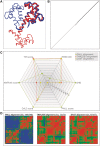
CSA is a web server for the computation, evaluation and comprehensive comparison of pairwise protein structure alignments. Its exact alignment engine computes either optimal, top-scoring alignments or heuristic alignments with quality guarantee for the inter-residue distance-based scorings of contact map overlap, PAUL, DALI and MATRAS. These and additional, uploaded alignments are compared using a number of quality measures and intuitive visualizations. CSA brings new insight into the structural relationship of the protein pairs under investigation and is a valuable tool for studying structural similarities. It is available at http://csa.project.cwi.nl.
Figures


References
-
- Hasegawa H, Holm L. Advances and pitfalls of protein structural alignment. Curr. Opin. Struct. Biol. 2009;19:341–348. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

