Signaling pathways in cell polarity
- PMID: 22553378
- PMCID: PMC3367552
- DOI: 10.1101/cshperspect.a009654
Signaling pathways in cell polarity
Abstract
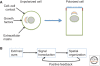
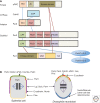
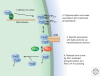
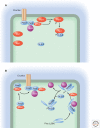
A key function of signal transduction during cell polarization is the creation of spatially segregated regions of the cell cortex that possess different lipid and protein compositions and have distinct functions. Polarity can be initiated spontaneously or in response to signaling inputs from adjacent cells or soluble factors and is stabilized by positive-feedback loops. A conserved group of proteins, the Par proteins, plays a central role in polarity establishment and maintenance in many contexts. These proteins generate and maintain their distinct locations in cells by actively excluding one another from specific regions of the plasma membrane. The Par signaling pathway intersects with multiple other pathways that control cell growth, death, and organization.
Figures

References
-
- Baas AF, Kuipers J, van der Wel NN, Batlle E, Koerten HK, Peters PJ, Clevers HC 2004. Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell 116: 457–466 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
