Lead identification and optimization of novel collagenase inhibitors; pharmacophore and structure based studies
- PMID: 22553386
- PMCID: PMC3338973
- DOI: 10.6026/97320630008301
Lead identification and optimization of novel collagenase inhibitors; pharmacophore and structure based studies
Abstract
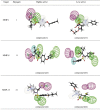
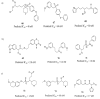
In this study, chemical feature based pharmacophore models of MMP-1, MMP-8 and MMP-13 inhibitors have been developed with the aid of HypoGen module within Catalyst program package. In MMP-1 and MMP-13, all the compounds in the training set mapped HBA and RA, while in MMP-8, the training set mapped HBA and HY. These features revealed responsibility for the high molecular bioactivity, and this is further used as a three dimensional query to screen the knowledge based designed molecules. These pharmacophore models for collagenases picked up some potent and novel inhibitors. Subsequently, docking studies were performed for the potent molecules and novel hits were suggested for further studies based on the docking score and active site interactions in MMP-1, MMP-8 and MMP-13.
Keywords: Collagenases; HypoGen; Induced Fit; Osteoarthritis; Pharmacophore; S1' loop.
Figures
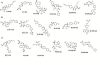
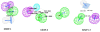
References
LinkOut - more resources
Full Text Sources
Miscellaneous
