Major Functional Transcriptome of an Inferred Center Regulator of an ER(-) Breast Cancer Model System
- PMID: 22553414
- PMCID: PMC3337785
- DOI: 10.4137/CIN.S8633
Major Functional Transcriptome of an Inferred Center Regulator of an ER(-) Breast Cancer Model System
Abstract
We aimed to find clinically relevant gene activities ruled by the signal transducer and activator of transcription 3 (STAT3) proteins in an ER(-) breast cancer population via network approach. STAT3 is negatively associated with both lymph nodal category and stage. MYC is a component of STAT3 network. MYC and STAT3 may co-regulate gene expressions for Warburg effect, stem cell like phenotype, cell proliferation and angiogenesis. We identified a STAT3 network in silico showing its ability in predicting its target gene expressions primarily for specific tumor subtype, tumor progression, treatment options and prognostic features. The aberrant expressions of MYC and STAT3 are enriched in triple negatives (TN). They promote histological grade, vascularity, metastasis and tumor anti-apoptotic activities. VEGFA, STAT3, FOXM1 and METAP2 are druggable targets. High levels of METAP2, MMP7, IGF2 and IGF2R are unfavorable prognostic factors. STAT3 is an inferred center regulator at early cancer development predominantly in TN.
Keywords: STAT3; grade; microarray; transcriptional regulatory network; vascularity.
Figures
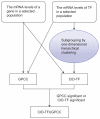
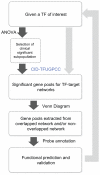


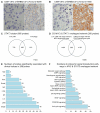
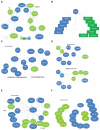

References
-
- Chen CJ, You SL, Lin LH, Hsu WL, Yang YW. Cancer epidemiology and control in Taiwan: a brief review. Jpn J Clin Oncol. 2002;32:S66–81. - PubMed
-
- Brown M, Bauer K, Pare M. Tumor marker phenotype concordance in second primary breast cancer, California, 1999–2004. Breast Cancer Res Treat. 2010;120:217–27. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

