Pattern analysis approach reveals restriction enzyme cutting abnormalities and other cDNA library construction artifacts using raw EST data
- PMID: 22554190
- PMCID: PMC3424822
- DOI: 10.1186/1472-6750-12-16
Pattern analysis approach reveals restriction enzyme cutting abnormalities and other cDNA library construction artifacts using raw EST data
Abstract
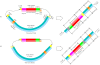
Background: Expressed Sequence Tag (EST) sequences are widely used in applications such as genome annotation, gene discovery and gene expression studies. However, some of GenBank dbEST sequences have proven to be "unclean". Identification of cDNA termini/ends and their structures in raw ESTs not only facilitates data quality control and accurate delineation of transcription ends, but also furthers our understanding of the potential sources of data abnormalities/errors present in the wet-lab procedures for cDNA library construction.
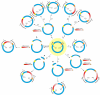
Results: After analyzing a total of 309,976 raw Pinus taeda ESTs, we uncovered many distinct variations of cDNA termini, some of which prove to be good indicators of wet-lab artifacts, and characterized each raw EST by its cDNA terminus structure patterns. In contrast to the expected patterns, many ESTs displayed complex and/or abnormal patterns that represent potential wet-lab errors such as: a failure of one or both of the restriction enzymes to cut the plasmid vector; a failure of the restriction enzymes to cut the vector at the correct positions; the insertion of two cDNA inserts into a single vector; the insertion of multiple and/or concatenated adapters/linkers; the presence of 3'-end terminal structures in designated 5'-end sequences or vice versa; and so on. With a close examination of these artifacts, many problematic ESTs that have been deposited into public databases by conventional bioinformatics pipelines or tools could be cleaned or filtered by our methodology. We developed a software tool for Abnormality Filtering and Sequence Trimming for ESTs (AFST, http://code.google.com/p/afst/) using a pattern analysis approach. To compare AFST with other pipelines that submitted ESTs into dbEST, we reprocessed 230,783 Pinus taeda and 38,709 Arachis hypogaea GenBank ESTs. We found 7.4% of Pinus taeda and 29.2% of Arachis hypogaea GenBank ESTs are "unclean" or abnormal, all of which could be cleaned or filtered by AFST.
Conclusions: cDNA terminal pattern analysis, as implemented in the AFST software tool, can be utilized to reveal wet-lab errors such as restriction enzyme cutting abnormities and chimeric EST sequences, detect various data abnormalities embedded in existing Sanger EST datasets, improve the accuracy of identifying and extracting bona fide cDNA inserts from raw ESTs, and therefore greatly benefit downstream EST-based applications.
Figures


References
-
- Cairney J, Zheng L, Cowels A, Hsiao J, Zismann V, Liu J, Ouyang S, Thibaud-Nissen F, Hamilton J, Childs K, Pullman GS, Zhang Y, Oh T, Buell CR. Expressed Sequence Tags from loblolly pine embryos reveal similarities with angiosperm embryogenesis. Plant Mol Biol. 2006;62:485–501. doi: 10.1007/s11103-006-9035-9. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

