Standardized representation, visualization and searchable repository of antiretroviral treatment-change episodes
- PMID: 22554313
- PMCID: PMC3439255
- DOI: 10.1186/1742-6405-9-13
Standardized representation, visualization and searchable repository of antiretroviral treatment-change episodes
Abstract
Background: To identify the determinants of successful antiretroviral (ARV) therapy, researchers study the virological responses to treatment-change episodes (TCEs) accompanied by baseline plasma HIV-1 RNA levels, CD4+ T lymphocyte counts, and genotypic resistance data. Such studies, however, often differ in their inclusion and virological response criteria making direct comparisons of study results problematic. Moreover, the absence of a standard method for representing the data comprising a TCE makes it difficult to apply uniform criteria in the analysis of published studies of TCEs.
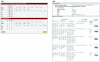
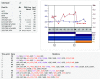
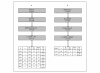
Results: To facilitate data sharing for TCE analyses, we developed an XML (Extensible Markup Language) Schema that represents the temporal relationship between plasma HIV-1 RNA levels, CD4 counts and genotypic drug resistance data surrounding an ARV treatment change. To demonstrate the adaptability of the TCE XML Schema to different clinical environments, we collaborate with four clinics to create a public repository of about 1,500 TCEs. Despite the nascent state of this TCE XML Repository, we were able to perform an analysis that generated a novel hypothesis pertaining to the optimal use of second-line therapies in resource-limited settings. We also developed an online program (TCE Finder) for searching the TCE XML Repository and another program (TCE Viewer) for generating a graphical depiction of a TCE from a TCE XML Schema document.
Conclusions: The TCE Suite of applications - the XML Schema, Viewer, Finder, and Repository - addresses several major needs in the analysis of the predictors of virological response to ARV therapy. The TCE XML Schema and Viewer facilitate sharing data comprising a TCE. The TCE Repository, the only publicly available collection of TCEs, and the TCE Finder can be used for testing the predictive value of genotypic resistance interpretation systems and potentially for generating and testing novel hypotheses pertaining to the optimal use of salvage ARV therapy.
Figures
Similar articles
-
XML Schema Representation of DICOM Structured Reporting.J Am Med Inform Assoc. 2003 Mar-Apr;10(2):213-23. doi: 10.1197/jamia.m1042. J Am Med Inform Assoc. 2003. PMID: 12595410 Free PMC article.
-
HIV-1 antiretroviral resistance: scientific principles and clinical applications.Drugs. 2012 Jun 18;72(9):e1-25. doi: 10.2165/11633630-000000000-00000. Drugs. 2012. PMID: 22686620 Free PMC article. Review.
-
Antiretroviral resistance testing in HIV-positive people.Cochrane Database Syst Rev. 2018 Nov 9;11(11):CD006495. doi: 10.1002/14651858.CD006495.pub5. Cochrane Database Syst Rev. 2018. PMID: 30411789 Free PMC article.
-
Development of a deterministic XML schema by resolving structure ambiguity of HL7 messages.Comput Methods Programs Biomed. 2005 Oct;80(1):1-15. doi: 10.1016/j.cmpb.2005.05.001. Comput Methods Programs Biomed. 2005. PMID: 15993979
-
Definition of an XML markup language for clinical laboratory procedures and comparison with generic XML markup.Clin Chem. 2006 Oct;52(10):1943-51. doi: 10.1373/clinchem.2006.071449. Epub 2006 Aug 3. Clin Chem. 2006. PMID: 16887897
Cited by
-
Using drug exposure for predicting drug resistance - A data-driven genotypic interpretation tool.PLoS One. 2017 Apr 10;12(4):e0174992. doi: 10.1371/journal.pone.0174992. eCollection 2017. PLoS One. 2017. PMID: 28394945 Free PMC article.
-
Determination of Phenotypic Resistance Cutoffs From Routine Clinical Data.J Acquir Immune Defic Syndr. 2017 Apr 15;74(5):e129-e137. doi: 10.1097/QAI.0000000000001198. J Acquir Immune Defic Syndr. 2017. PMID: 27787339 Free PMC article.
References
-
- Brun-Vezinet F, Costagliola D, Khaled MA, Calvez V, Clavel F, Clotet B, Haubrich R, Kempf D, King M, Kuritzkes D. et al.Clinically validated genotype analysis: guiding principles and statistical concerns. Antivir Ther. 2004;9:465–478. - PubMed
-
- Kempf DJ, Isaacson JD, King MS, Brun SC, Sylte J, Richards B, Bernstein B, Rode R, Sun E. Analysis of the virological response with respect to baseline viral phenotype and genotype in protease inhibitor-experienced HIV-1-infected patients receiving lopinavir/ritonavir therapy. Antivir Ther. 2002;7:165–174. - PubMed
-
- Lanier ER, Ait-Khaled M, Scott J, Stone C, Melby T, Sturge G, St Clair M, Steel H, Hetherington S, Pearce G. et al.Antiviral efficacy of abacavir in antiretroviral therapy-experienced adults harbouring HIV-1 with specific patterns of resistance to nucleoside reverse transcriptase inhibitors. Antivir Ther. 2004;9:37–45. - PubMed
-
- Baxter JD, Schapiro JM, Boucher CA, Kohlbrenner VM, Hall DB, Scherer JR, Mayers DL. Genotypic changes in human immunodeficiency virus type 1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. J Virol. 2006;80:10794–10801. doi: 10.1128/JVI.00712-06. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

