The specialized unfolded protein response of B lymphocytes: ATF6α-independent development of antibody-secreting B cells
- PMID: 22555069
- PMCID: PMC3358488
- DOI: 10.1016/j.molimm.2012.04.001
The specialized unfolded protein response of B lymphocytes: ATF6α-independent development of antibody-secreting B cells
Abstract
B lymphocytes, like all mammalian cells, are equipped with the unfolded protein response (UPR), a complex signaling system allowing for both pro- and mal-adaptive responses to increased demands on the endoplasmic reticulum (ER). The UPR is comprised of three signaling pathways initiated by the ER transmembrane stress sensors, IRE1α/β, PERK and ATF6α/β. Activation of IRE1 yields XBP1(S), a transcription factor that directs expansion of the ER and enhances protein biosynthetic and secretory machinery. XBP1(S) is essential for the differentiation of B lymphocytes into antibody-secreting cells. In contrast, the PERK pathway, a regulator of translation and transcription, is dispensable for the generation of antibody-secreting cells. Functioning as a transcription factor, ATF6α can augment ER quality control processes and drive ER expansion, but the potential role of this UPR pathway in activated B cells has not been investigated. Here, we report studies of ATF6α-deficient B cells demonstrating that ATF6α is not required for the development of antibody-secreting cells. Thus, when B cells are stimulated to secrete antibody, a specialized UPR relies exclusively on the IRE1-XBP1 pathway to remodel the ER and expand cellular secretory capacity.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
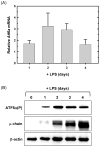




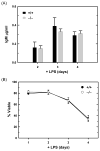

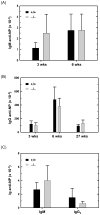
References
-
- Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. - PubMed
-
- Brunsing R, Omori SA, Weber F, Bicknell A, Friend L, Rickert R, Niwa M. B- and T-cell development both involve activity of the unfolded protein response pathway. J Biol Chem. 2008;283:17954–17961. - PubMed
-
- Calame KL, Lin KI, Tunyaplin C. Regulatory mechanisms that determine the development and function of plasma cells. Annu Rev Immunol. 2003;21:205–230. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

