Genome-wide copy number analysis of single cells
- PMID: 22555242
- PMCID: PMC5069701
- DOI: 10.1038/nprot.2012.039
Genome-wide copy number analysis of single cells
Erratum in
-
Corrigendum: Genome-wide copy number analysis of single cells.Nat Protoc. 2016 Mar;11(3):616. doi: 10.1038/nprot0316.616b. Epub 2016 Feb 25. Nat Protoc. 2016. PMID: 26914320 No abstract available.
Abstract
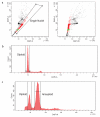
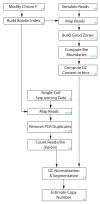
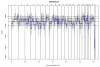
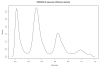
Copy number variation (CNV) is increasingly recognized as an important contributor to phenotypic variation in health and disease. Most methods for determining CNV rely on admixtures of cells in which information regarding genetic heterogeneity is lost. Here we present a protocol that allows for the genome-wide copy number analysis of single nuclei isolated from mixed populations of cells. Single-nucleus sequencing (SNS), combines flow sorting of single nuclei on the basis of DNA content and whole-genome amplification (WGA); this is followed by next-generation sequencing to quantize genomic intervals in a genome-wide manner. Multiplexing of single cells is discussed. In addition, we outline informatic approaches that correct for biases inherent in the WGA procedure and allow for accurate determination of copy number profiles. All together, the protocol takes ∼3 d from flow cytometry to sequence-ready DNA libraries.
Figures





References
-
- Beckmann JS, Estivill X, Antonarakis SE. Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nat Rev Genet. 2007;8:639–646. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

