Cellular function and molecular structure of ecto-nucleotidases
- PMID: 22555564
- PMCID: PMC3360096
- DOI: 10.1007/s11302-012-9309-4
Cellular function and molecular structure of ecto-nucleotidases
Abstract
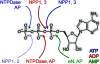
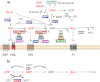
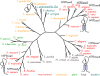
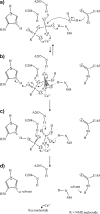
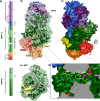
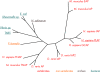
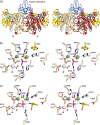
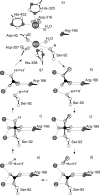
Ecto-nucleotidases play a pivotal role in purinergic signal transmission. They hydrolyze extracellular nucleotides and thus can control their availability at purinergic P2 receptors. They generate extracellular nucleosides for cellular reuptake and salvage via nucleoside transporters of the plasma membrane. The extracellular adenosine formed acts as an agonist of purinergic P1 receptors. They also can produce and hydrolyze extracellular inorganic pyrophosphate that is of major relevance in the control of bone mineralization. This review discusses and compares four major groups of ecto-nucleotidases: the ecto-nucleoside triphosphate diphosphohydrolases, ecto-5'-nucleotidase, ecto-nucleotide pyrophosphatase/phosphodiesterases, and alkaline phosphatases. Only recently and based on crystal structures, detailed information regarding the spatial structures and catalytic mechanisms has become available for members of these four ecto-nucleotidase families. This permits detailed predictions of their catalytic mechanisms and a comparison between the individual enzyme groups. The review focuses on the principal biochemical, cell biological, catalytic, and structural properties of the enzymes and provides brief reference to tissue distribution, and physiological and pathophysiological functions.
Figures



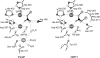
References
-
- Zimmermann H, Mishra SK, Shukla V, Langer D, Gampe K, Grimm I, Delic J, Braun N. Ecto-nucleotidases, molecular properties and functional impact. An R Acad Nac Farm. 2007;73:537–566.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

