Looking deep inside: detection of low-abundance proteins in leaf extracts of Arabidopsis and phloem exudates of pumpkin
- PMID: 22555880
- PMCID: PMC3387715
- DOI: 10.1104/pp.112.198077
Looking deep inside: detection of low-abundance proteins in leaf extracts of Arabidopsis and phloem exudates of pumpkin
Abstract
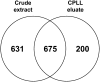
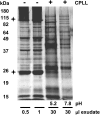
The field of proteomics suffers from the immense complexity of even small proteomes and the enormous dynamic range of protein concentrations within a given sample. Most protein samples contain a few major proteins, which hamper in-depth proteomic analysis. In the human field, combinatorial hexapeptide ligand libraries (CPLL; such as ProteoMiner) have been used for reduction of the dynamic range of protein concentrations; however, this technique is not established in plant research. In this work, we present the application of CPLL to Arabidopsis (Arabidopsis thaliana) leaf proteins. One- and two-dimensional gel electrophoresis showed a decrease in high-abundance proteins and an enrichment of less abundant proteins in CPLL-treated samples. After optimization of the CPLL protocol, mass spectrometric analyses of leaf extracts led to the identification of 1,192 proteins in control samples and an additional 512 proteins after the application of CPLL. Upon leaf infection with virulent Pseudomonas syringae DC3000, CPLL beads were also used for investigating the bacterial infectome. In total, 312 bacterial proteins could be identified in infected Arabidopsis leaves. Furthermore, phloem exudates of pumpkin (Cucurbita maxima) were analyzed. CPLL prefractionation caused depletion of the major phloem proteins 1 and 2 and improved phloem proteomics, because 67 of 320 identified proteins were detectable only after CPLL treatment. In sum, our results demonstrate that CPLL beads are a time- and cost-effective tool for reducing major proteins, which often interfere with downstream analyses. The concomitant enrichment of less abundant proteins may facilitate a deeper insight into the plant proteome.
Figures





References
-
- Ardales E, Moon S-J, Park DS, Sr, Byun M-O, Noh TH. (2009) Inactivation of argG, encoding argininosuccinate synthetase from Xanthomonas oryzae pv. oryzae, is involved in bacterial growth and virulence in planta. Can J Plant Pathol 31: 368–374
-
- Atkins CA, Smith PM, Rodriguez-Medina C. (2011) Macromolecules in phloem exudates: a review. Protoplasma 248: 165–172 - PubMed
-
- Bachi A, Simó C, Restuccia U, Guerrier L, Fortis F, Boschetti E, Masseroli M, Righetti PG. (2008) Performance of combinatorial peptide libraries in capturing the low-abundance proteome of red blood cells. 2. Behavior of resins containing individual amino acids. Anal Chem 80: 3557–3565 - PubMed
-
- Boschetti E, Lomas L, Citterio A, Righetti PG. (2007) Romancing the “hidden proteome,” Anno Domini two zero zero seven. J Chromatogr A 1153: 277–290 - PubMed
-
- Boschetti E, Righetti PG. (2008a) Hexapeptide combinatorial ligand libraries: the march for the detection of the low-abundance proteome continues. Biotechniques 44: 663–665 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

