SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes
- PMID: 22556368
- PMCID: PMC3389763
- DOI: 10.1093/bioinformatics/bts252
SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes
Abstract
Motivation: In the analysis of homologous sequences, computation of multiple sequence alignments (MSAs) has become a bottleneck. This is especially troublesome for marker genes like the ribosomal RNA (rRNA) where already millions of sequences are publicly available and individual studies can easily produce hundreds of thousands of new sequences. Methods have been developed to cope with such numbers, but further improvements are needed to meet accuracy requirements.
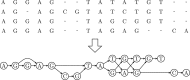
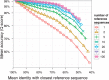
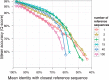
Results: In this study, we present the SILVA Incremental Aligner (SINA) used to align the rRNA gene databases provided by the SILVA ribosomal RNA project. SINA uses a combination of k-mer searching and partial order alignment (POA) to maintain very high alignment accuracy while satisfying high throughput performance demands. SINA was evaluated in comparison with the commonly used high throughput MSA programs PyNAST and mothur. The three BRAliBase III benchmark MSAs could be reproduced with 99.3, 97.6 and 96.1 accuracy. A larger benchmark MSA comprising 38 772 sequences could be reproduced with 98.9 and 99.3% accuracy using reference MSAs comprising 1000 and 5000 sequences. SINA was able to achieve higher accuracy than PyNAST and mothur in all performed benchmarks.
Availability: Alignment of up to 500 sequences using the latest SILVA SSU/LSU Ref datasets as reference MSA is offered at http://www.arb-silva.de/aligner. This page also links to Linux binaries, user manual and tutorial. SINA is made available under a personal use license.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

