Ss-Sl2, a novel cell wall protein with PAN modules, is essential for sclerotial development and cellular integrity of Sclerotinia sclerotiorum
- PMID: 22558105
- PMCID: PMC3338822
- DOI: 10.1371/journal.pone.0034962
Ss-Sl2, a novel cell wall protein with PAN modules, is essential for sclerotial development and cellular integrity of Sclerotinia sclerotiorum
Abstract
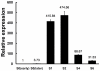
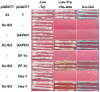
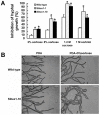
The sclerotium is an important dormant body for many plant fungal pathogens. Here, we reported that a protein, named Ss-Sl2, is involved in sclerotial development of Sclerotinia sclerotiorum. Ss-Sl2 does not show significant homology with any protein of known function. Ss-Sl2 contains two putative PAN modules which were found in other proteins with diverse adhesion functions. Ss-Sl2 is a secreted protein, during the initial stage of sclerotial development, copious amounts of Ss-Sl2 are secreted and accumulated on the cell walls. The ability to maintain the cellular integrity of RNAi-mediated Ss-Sl2 silenced strains was reduced, but the hyphal growth and virulence of Ss-Sl2 silenced strains were not significantly different from the wild strain. Ss-Sl2 silenced strains could form interwoven hyphal masses at the initial stage of sclerotial development, but the interwoven hyphae could not consolidate and melanize. Hyphae in these interwoven bodies were thin-walled, and arranged loosely. Co-immunoprecipitation and yeast two-hybrid experiments showed that glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Woronin body major protein (Hex1) and elongation factor 1-alpha interact with Ss-Sl2. GAPDH-knockdown strains showed a similar phenotype in sclerotial development as Ss-Sl2 silenced strains. Hex1-knockdown strains showed similar impairment in maintenance of hyphal integrity as Ss-Sl2 silenced strains. The results suggested that Ss-Sl2 functions in both sclerotial development and cellular integrity of S. sclerotiorum.
Conflict of interest statement
Figures









Similar articles
-
Ss-Rhs1, a secretory Rhs repeat-containing protein, is required for the virulence of Sclerotinia sclerotiorum.Mol Plant Pathol. 2017 Oct;18(8):1052-1061. doi: 10.1111/mpp.12459. Epub 2016 Sep 8. Mol Plant Pathol. 2017. PMID: 27392818 Free PMC article.
-
An atypical forkhead-containing transcription factor SsFKH1 is involved in sclerotial formation and is essential for pathogenicity in Sclerotinia sclerotiorum.Mol Plant Pathol. 2017 Sep;18(7):963-975. doi: 10.1111/mpp.12453. Epub 2016 Aug 21. Mol Plant Pathol. 2017. PMID: 27353472 Free PMC article.
-
RNA interference-mediated targeting of monooxygenase SsMNO1 for controlling Sclerotinia stem rot caused by Sclerotinia sclerotiorum.Pest Manag Sci. 2025 Mar;81(3):1457-1468. doi: 10.1002/ps.8546. Epub 2024 Nov 18. Pest Manag Sci. 2025. PMID: 39555684
-
Proteomic Analysis Reveals the Importance of Exudates on Sclerotial Development in Sclerotinia sclerotiorum.J Agric Food Chem. 2021 Feb 3;69(4):1430-1440. doi: 10.1021/acs.jafc.0c06685. Epub 2021 Jan 22. J Agric Food Chem. 2021. PMID: 33481591
-
Characterization of the Sclerotinia sclerotiorum cell wall proteome.Mol Plant Pathol. 2016 Aug;17(6):985-95. doi: 10.1111/mpp.12352. Epub 2016 Apr 4. Mol Plant Pathol. 2016. PMID: 26661933 Free PMC article.
Cited by
-
A secretory protein of necrotrophic fungus Sclerotinia sclerotiorum that suppresses host resistance.PLoS One. 2013;8(1):e53901. doi: 10.1371/journal.pone.0053901. Epub 2013 Jan 14. PLoS One. 2013. PMID: 23342034 Free PMC article.
-
Two distinct classes of protein related to GTB and RRM are critical in the sclerotial metamorphosis process of Rhizoctonia solani AG-1 IA.Funct Integr Genomics. 2015 Jul;15(4):449-59. doi: 10.1007/s10142-015-0435-2. Epub 2015 Mar 13. Funct Integr Genomics. 2015. PMID: 25763752
-
Characterization of Transcriptional Responses to Genomovirus Infection of the White Mold Fungus, Sclerotinia sclerotiorum.Viruses. 2022 Aug 27;14(9):1892. doi: 10.3390/v14091892. Viruses. 2022. PMID: 36146699 Free PMC article.
-
An effector SsCVNH promotes the virulence of Sclerotinia sclerotiorum through targeting class III peroxidase AtPRX71.Mol Plant Pathol. 2024 May;25(5):e13464. doi: 10.1111/mpp.13464. Mol Plant Pathol. 2024. PMID: 38695733 Free PMC article.
-
Comparative genomic and transcriptional analyses of the carbohydrate-active enzymes and secretomes of phytopathogenic fungi reveal their significant roles during infection and development.Sci Rep. 2015 Nov 4;5:15565. doi: 10.1038/srep15565. Sci Rep. 2015. PMID: 26531059 Free PMC article.
References
-
- Boland GJ, Hall R. Index of plant hosts of Sclerotinia sclerotiorum. Can J Plant Pathol. 1994;16:93–100.
-
- Bolton M, Thomma BPHJ, Nelson B. Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol Plant Pathol. 2006;7:1–16. - PubMed
-
- Steadman JR. Control of plant diseases caused by Sclerotinia species. Phytopathology. 1979;69:904–907.
-
- Kosasih BD, Willetts HJ. Ontogenetic and histochemical studies of the apothecium of Sclerotinia sclerotiorum. Ann Bot. 1975;39:185–191.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

