Candidate proteins, metabolites and transcripts in the Biomarkers for Spinal Muscular Atrophy (BforSMA) clinical study
- PMID: 22558154
- PMCID: PMC3338723
- DOI: 10.1371/journal.pone.0035462
Candidate proteins, metabolites and transcripts in the Biomarkers for Spinal Muscular Atrophy (BforSMA) clinical study
Abstract
Background: Spinal Muscular Atrophy (SMA) is a neurodegenerative motor neuron disorder resulting from a homozygous mutation of the survival of motor neuron 1 (SMN1) gene. The gene product, SMN protein, functions in RNA biosynthesis in all tissues. In humans, a nearly identical gene, SMN2, rescues an otherwise lethal phenotype by producing a small amount of full-length SMN protein. SMN2 copy number inversely correlates with disease severity. Identifying other novel biomarkers could inform clinical trial design and identify novel therapeutic targets.
Objective: To identify novel candidate biomarkers associated with disease severity in SMA using unbiased proteomic, metabolomic and transcriptomic approaches.
Materials and methods: A cross-sectional single evaluation was performed in 108 children with genetically confirmed SMA, aged 2-12 years, manifesting a broad range of disease severity and selected to distinguish factors associated with SMA type and present functional ability independent of age. Blood and urine specimens from these and 22 age-matched healthy controls were interrogated using proteomic, metabolomic and transcriptomic discovery platforms. Analyte associations were evaluated against a primary measure of disease severity, the Modified Hammersmith Functional Motor Scale (MHFMS) and to a number of secondary clinical measures.
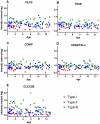
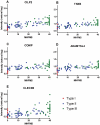
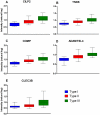
Results: A total of 200 candidate biomarkers correlate with MHFMS scores: 97 plasma proteins, 59 plasma metabolites (9 amino acids, 10 free fatty acids, 12 lipids and 28 GC/MS metabolites) and 44 urine metabolites. No transcripts correlated with MHFMS.
Discussion: In this cross-sectional study, "BforSMA" (Biomarkers for SMA), candidate protein and metabolite markers were identified. No transcript biomarker candidates were identified. Additional mining of this rich dataset may yield important insights into relevant SMA-related pathophysiology and biological network associations. Additional prospective studies are needed to confirm these findings, demonstrate sensitivity to change with disease progression, and assess potential impact on clinical trial design.
Trial registry: Clinicaltrials.gov NCT00756821.
Conflict of interest statement
Figures
References
-
- Mattis VB, Rai R, Wang J, Chang CW, Coady T, et al. Novel aminoglycosides increase SMN levels in spinal muscular atrophy fibroblasts. Hum Genet. 2006;120:589–601. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- ARRA 5-R01 HD054599-04/HD/NICHD NIH HHS/United States
- 1R21 AI078387-S1/AI/NIAID NIH HHS/United States
- UL1-RR 025005/RR/NCRR NIH HHS/United States
- UL1 RR024992/RR/NCRR NIH HHS/United States
- 1R21, AR059466/AR/NIAMS NIH HHS/United States
- 1R41 AI071927/AI/NIAID NIH HHS/United States
- 1 NIH UL1-RR-024134/RR/NCRR NIH HHS/United States
- AR058606/AR/NIAMS NIH HHS/United States
- UL1-RR025764/RR/NCRR NIH HHS/United States
- UL1 RR025005/RR/NCRR NIH HHS/United States
- UL1 RR025764/RR/NCRR NIH HHS/United States
- 1-R01-HD69045/HD/NICHD NIH HHS/United States
- T32, AR059650/AR/NIAMS NIH HHS/United States
- UL1 RR025780/RR/NCRR NIH HHS/United States
- UL1-RR025755/RR/NCRR NIH HHS/United States
- UL1 RR024156/RR/NCRR NIH HHS/United States
- M01 RR002172/RR/NCRR NIH HHS/United States
- 1UL1-RR024979/RR/NCRR NIH HHS/United States
- R01-HD054599/HD/NICHD NIH HHS/United States
- KL2 RR024980/RR/NCRR NIH HHS/United States
- UL1 RR025758-01/RR/NCRR NIH HHS/United States
- 1U19AI082726, T32/AI/NIAID NIH HHS/United States
- 1R21 AI078387/AI/NIAID NIH HHS/United States
- C06 RR011234/RR/NCRR NIH HHS/United States
- UL1 RR025755/RR/NCRR NIH HHS/United States
- UL1-RR024992/RR/NCRR NIH HHS/United States
- UL1RR025780/RR/NCRR NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- UL1 RR025758/RR/NCRR NIH HHS/United States
- 1TL1-RR024981/RR/NCRR NIH HHS/United States
- NIH 1 UL1 RR024156/RR/NCRR NIH HHS/United States
- MO1-RR02172/RR/NCRR NIH HHS/United States
- TL1 RR024981/RR/NCRR NIH HHS/United States
- R01 AI063623/AI/NIAID NIH HHS/United States
- 1KL2-RR024980/RR/NCRR NIH HHS/United States
- UL1 RR024134/RR/NCRR NIH HHS/United States
- UL1 RR024979/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

