In vitro RNase and nucleic acid binding activities implicate coilin in U snRNA processing
- PMID: 22558428
- PMCID: PMC3338655
- DOI: 10.1371/journal.pone.0036300
In vitro RNase and nucleic acid binding activities implicate coilin in U snRNA processing
Abstract
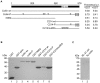
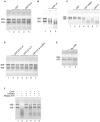
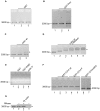
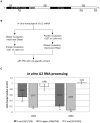
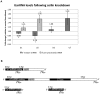
Coilin is known as the marker protein for Cajal bodies (CBs), subnuclear domains important for the biogenesis of small nuclear ribonucleoproteins (snRNPs) which function in pre-mRNA splicing. CBs associate non-randomly with U1 and U2 gene loci, which produce the small nuclear RNA (snRNA) component of the respective snRNP. Despite recognition as the CB marker protein, coilin is primarily nucleoplasmic, and the function of this fraction is not fully characterized. Here we show that coilin binds double stranded DNA and has RNase activity in vitro. U1 and U2 snRNAs undergo a processing event of the primary transcript prior to incorporation in the snRNP. We find that coilin displays RNase activity within the CU region of the U2 snRNA primary transcript in vitro, and that coilin knockdown results in accumulation of the 3' pre-processed U1 and U2 snRNA. These findings present new characteristics of coilin in vitro, and suggest additional functions of the protein in vivo.
Conflict of interest statement
Figures
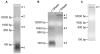
References
-
- Morris GE. The Cajal body. Biochim Biophys Acta. 2008;1783:2108–2115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

