Label-free quantitative LC-MS proteomics of Alzheimer's disease and normally aged human brains
- PMID: 22559202
- PMCID: PMC3445701
- DOI: 10.1021/pr3001546
Label-free quantitative LC-MS proteomics of Alzheimer's disease and normally aged human brains
Abstract
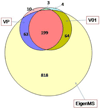
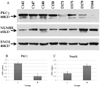
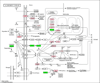
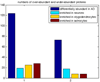
Quantitative proteomics analysis of cortical samples of 10 Alzheimer's disease (AD) brains versus 10 normally aged brains was performed by following the accurate mass and time tag (AMT) approach with the high resolution LTQ Orbitrap mass spectrometer. More than 1400 proteins were identified and quantitated. A conservative approach of selecting only the consensus results of four normalization methods was suggested and used. A total of 197 proteins were shown to be significantly differentially abundant (p-values <0.05, corrected for multiplicity of testing) in AD versus control brain samples. Thirty-seven of these proteins were reported as differentially abundant or modified in AD in previous proteomics and transcriptomics publications. The rest to the best of our knowledge are new. Mapping of the discovered proteins with bioinformatic tools revealed significant enrichment with differentially abundant proteins of pathways and processes known to be important in AD, including signal transduction, regulation of protein phosphorylation, immune response, cytoskeleton organization, lipid metabolism, energy production, and cell death.
Conflict of interest statement
Authors declare no conflict of interest.
Figures



References
-
- Evans DA, Funkenstein HH, Albert MS, Scherr PA, Cook NR, Chown MJ, Hebert LE, Hennekens CH, Taylor JO. Prevalence of Alzheimer’s disease in a community population of older persons. Higher than previously reported. JAMA. 1989;262:2551–2556. - PubMed
-
- Gatz M, Pedersen NL, Berg S, Johansson B, Johansson K, Mortimer JA, Posner SF, Viitanen M, Winblad B, Ahlbom A. Heritability for Alzheimer’s disease: the study of dementia in Swedish twins. J Gerontol A Biol Sci Med Sci. 1997;52:M117–M125. - PubMed
-
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. - PubMed
-
- Lovestone S, Guntert A, Hye A, Lynham S, Thambisetty M, Ward M. Proteomics of Alzheimer’s disease: understanding mechanisms and seeking biomarkers. Expert Rev Proteomics. 2007;4:227–238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

