Optical nanoscopy: from acquisition to analysis
- PMID: 22559319
- PMCID: PMC3568761
- DOI: 10.1146/annurev-bioeng-071811-150025
Optical nanoscopy: from acquisition to analysis
Abstract
Recent advances in far-field microscopy have demonstrated that fluorescence imaging is possible at resolutions well below the long-standing diffraction limit. By exploiting photophysical properties of fluorescent probe molecules, this new class of methods yields a resolving power that is fundamentally diffraction unlimited. Although these methods are becoming more widely used in biological imaging, they must be complemented by suitable data analysis approaches if their potential is to be fully realized. Here we review the basic principles of diffraction-unlimited microscopy and how these principles influence the selection of available algorithms for data analysis. Furthermore, we provide an overview of existing analysis strategies and discuss their application.
Conflict of interest statement
The authors hold patents in the field and have financial interest in Vutara, Inc.
Figures
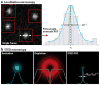

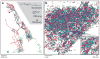

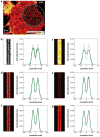
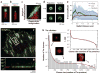
References
-
- Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–58. - PubMed
-
- Toomre D, Bewersdorf J. A new wave of cellular imaging. Annu Rev Cell Dev Biol. 2010;26:285–314. - PubMed
-
- Abbe E. Beiträge zur Theorie des Mikroskops und der Mikroskopischen Wahrnehmung. Arch Mikrosk Anat. 1873;9:413–68.
-
- Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–82. - PubMed
-
- Hell SW. Microscopy and its focal switch. Nat Methods. 2009;6:24–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

