Identification of genes with a correlation between copy number and expression in gastric cancer
- PMID: 22559327
- PMCID: PMC3441862
- DOI: 10.1186/1755-8794-5-14
Identification of genes with a correlation between copy number and expression in gastric cancer
Abstract
Background: To elucidate gene expression associated with copy number changes, we performed a genome-wide copy number and expression microarray analysis of 25 pairs of gastric tissues.
Methods: We applied laser capture microdissection (LCM) to obtain samples for microarray experiments and profiled DNA copy number and gene expression using 244K CGH Microarray and Human Exon 1.0 ST Microarray.
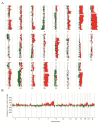
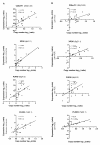
Results: Obviously, gain at 8q was detected at the highest frequency (70%) and 20q at the second (63%). We also identified molecular genetic divergences for different TNM-stages or histological subtypes of gastric cancers. Interestingly, the C20orf11 amplification and gain at 20q13.33 almost separated moderately differentiated (MD) gastric cancers from poorly differentiated (PD) type. A set of 163 genes showing the correlations between gene copy number and expression was selected and the identified genes were able to discriminate matched adjacent noncancerous samples from gastric cancer samples in an unsupervised two-way hierarchical clustering. Quantitative RT-PCR analysis for 4 genes (C20orf11, XPO5, PUF60, and PLOD3) of the 163 genes validated the microarray results. Notably, some candidate genes (MCM4 and YWHAZ) and its adjacent genes such as PRKDC, UBE2V2, ANKRD46, ZNF706, and GRHL2, were concordantly deregulated by genomic aberrations.
Conclusions: Taken together, our results reveal diverse chromosomal region alterations for different TNM-stages or histological subtypes of gastric cancers, which is helpful in researching clinicopathological classification, and highlight several interesting genes as potential biomarkers for gastric cancer.
Figures

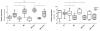


Similar articles
-
Genome-wide analysis of DNA copy number alterations and gene expression in gastric cancer.J Pathol. 2008 Dec;216(4):471-82. doi: 10.1002/path.2424. J Pathol. 2008. PMID: 18798223
-
High resolution analysis of DNA copy-number aberrations of chromosomes 8, 13, and 20 in gastric cancers.Virchows Arch. 2009 Sep;455(3):213-23. doi: 10.1007/s00428-009-0814-y. Epub 2009 Aug 21. Virchows Arch. 2009. PMID: 19697059 Free PMC article.
-
Integration of DNA copy number alterations and transcriptional expression analysis in human gastric cancer.PLoS One. 2012;7(4):e29824. doi: 10.1371/journal.pone.0029824. Epub 2012 Apr 23. PLoS One. 2012. PMID: 22539939 Free PMC article.
-
Copy number alterations and expression profiles of candidate genes in a pulmonary inflammatory myofibroblastic tumor.Lung Cancer. 2010 Nov;70(2):152-7. doi: 10.1016/j.lungcan.2010.01.019. Epub 2010 Feb 24. Lung Cancer. 2010. PMID: 20185201 Review.
-
Search for new biomarkers of gastric cancer through serial analysis of gene expression and its clinical implications.Cancer Sci. 2004 May;95(5):385-92. doi: 10.1111/j.1349-7006.2004.tb03220.x. Cancer Sci. 2004. PMID: 15132764 Free PMC article. Review.
Cited by
-
Integrating biological knowledge and gene expression data using pathway-guided random forests: a benchmarking study.Bioinformatics. 2020 Aug 1;36(15):4301-4308. doi: 10.1093/bioinformatics/btaa483. Bioinformatics. 2020. PMID: 32399562 Free PMC article.
-
Grainyhead-like 2 as a double-edged sword in development and cancer.Am J Transl Res. 2020 Feb 15;12(2):310-331. eCollection 2020. Am J Transl Res. 2020. PMID: 32194886 Free PMC article. Review.
-
Exploration and Clinical Verification of the Blood Co-Expression Genes of Type 2 Diabetes Mellitus and Mild Cognitive Dysfunction in the Elderly.Biomedicines. 2023 Mar 23;11(4):993. doi: 10.3390/biomedicines11040993. Biomedicines. 2023. PMID: 37189611 Free PMC article.
-
PUF60 Promotes Chemoresistance Through Drug Efflux and Reducing Apoptosis in Gastric Cancer.Int J Med Sci. 2025 Jan 1;22(2):269-282. doi: 10.7150/ijms.102976. eCollection 2025. Int J Med Sci. 2025. PMID: 39781520 Free PMC article.
-
Overexpression of MAPK15 in gastric cancer is associated with copy number gain and contributes to the stability of c-Jun.Oncotarget. 2015 Aug 21;6(24):20190-203. doi: 10.18632/oncotarget.4171. Oncotarget. 2015. PMID: 26035356 Free PMC article.
References
-
- Powell SM. In: In the Genetic Basis of Human Cancer. 2. Vogelstein B, Kinzler K, editor. McGraw-Hill, New York; 2002. Stomach cancer; pp. 703–708.
-
- Tahara E. Oncogenes in human gastric carcinoma. Gan To Kagaku Ryoho. 1989;16(6):2149–2155. - PubMed
-
- Jinawath N, Furukawa Y, Hasegawa S, Li M, Tsunoda T, Satoh S, Yamaguchi T, Imamura H, Inoue M, Shiozaki H. et al.Comparison of gene-expression profiles between diffuse- and intestinal-type gastric cancers using a genome-wide cDNA microarray. Oncogene. 2004;23(40):6830–6844. doi: 10.1038/sj.onc.1207886. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

