NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment
- PMID: 22560079
- PMCID: PMC3402183
- DOI: 10.1016/j.stem.2012.02.020
NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment
Abstract
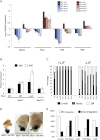
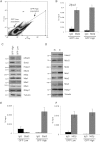
Transcriptional heterogeneity within embryonic stem cell (ESC) populations has been suggested as a mechanism by which a seemingly homogeneous cell population can initiate differentiation into an array of different cell types. Chromatin remodeling proteins have been shown to control transcriptional variability in yeast and to be important for mammalian ESC lineage commitment. Here we show that the Nucleosome Remodeling and Deacetylation (NuRD) complex, which is required for ESC lineage commitment, modulates both transcriptional heterogeneity and the dynamic range of a set of pluripotency genes in ESCs. In self-renewing conditions, the influence of NuRD at these genes is balanced by the opposing action of self-renewal factors. Upon loss of self-renewal factors, the action of NuRD is sufficient to silence transcription of these pluripotency genes, allowing cells to exit self-renewal. We propose that modulation of transcription levels by NuRD is key to maintaining the differentiation responsiveness of pluripotent cells.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures






References
-
- Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. - PubMed
-
- Chambers I. The molecular basis of pluripotency in mouse embryonic stem cells. Cloning Stem Cells. 2004;6:386–391. - PubMed
-
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. - PubMed
-
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. - PubMed
-
- Chen C., Sun X., Ran Q., Wilkinson K.D., Murphy T.J., Simons J.W., Dong J.-T. Ubiquitin-proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene. 2005;24:3319–3327. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

