A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits
- PMID: 22560090
- PMCID: PMC3376551
- DOI: 10.1016/j.ajhg.2012.03.015
A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits
Abstract
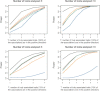
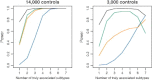
Pooling genome-wide association studies (GWASs) increases power but also poses methodological challenges because studies are often heterogeneous. For example, combining GWASs of related but distinct traits can provide promising directions for the discovery of loci with small but common pleiotropic effects. Classical approaches for meta-analysis or pooled analysis, however, might not be suitable for such analysis because individual variants are likely to be associated with only a subset of the traits or might demonstrate effects in different directions. We propose a method that exhaustively explores subsets of studies for the presence of true association signals that are in either the same direction or possibly opposite directions. An efficient approximation is used for rapid evaluation of p values. We present two illustrative applications, one for a meta-analysis of separate case-control studies of six distinct cancers and another for pooled analysis of a case-control study of glioma, a class of brain tumors that contains heterogeneous subtypes. Both the applications and additional simulation studies demonstrate that the proposed methods offer improved power and more interpretable results when compared to traditional methods for the analysis of heterogeneous traits. The proposed framework has applications beyond genetic association studies.
Copyright © 2012 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

