A human homeotic transformation resulting from mutations in PLCB4 and GNAI3 causes auriculocondylar syndrome
- PMID: 22560091
- PMCID: PMC3376493
- DOI: 10.1016/j.ajhg.2012.04.002
A human homeotic transformation resulting from mutations in PLCB4 and GNAI3 causes auriculocondylar syndrome
Erratum in
- Am J Hum Genet. 2012 Aug 10;91(2):397
- Am J Hum Genet. 2012 Jun 8;90(6):1116
Abstract
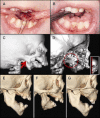
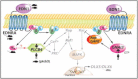
Auriculocondylar syndrome (ACS) is a rare, autosomal-dominant craniofacial malformation syndrome characterized by variable micrognathia, temporomandibular joint ankylosis, cleft palate, and a characteristic "question-mark" ear malformation. Careful phenotypic characterization of severely affected probands in our cohort suggested the presence of a mandibular patterning defect resulting in a maxillary phenotype (i.e., homeotic transformation). We used exome sequencing of five probands and identified two novel (exclusive to the patient and/or family studied) missense mutations in PLCB4 and a shared mutation in GNAI3 in two unrelated probands. In confirmatory studies, three additional novel PLCB4 mutations were found in multigenerational ACS pedigrees. All mutations were confirmed by Sanger sequencing, were not present in more than 10,000 control chromosomes, and resulted in amino-acid substitutions located in highly conserved protein domains. Additionally, protein-structure modeling demonstrated that all ACS substitutions disrupt the catalytic sites of PLCB4 and GNAI3. We suggest that PLCB4 and GNAI3 are core signaling molecules of the endothelin-1-distal-less homeobox 5 and 6 (EDN1-DLX5/DLX6) pathway. Functional studies demonstrated a significant reduction in downstream DLX5 and DLX6 expression in ACS cases in assays using cultured osteoblasts from probands and controls. These results support the role of the previously implicated EDN1-DLX5/6 pathway in regulating mandibular specification in other species, which, when disrupted, results in a maxillary phenotype. This work defines the molecular basis of ACS as a homeotic transformation (mandible to maxilla) in humans.
Copyright © 2012 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Uuspää V. Combined bilateral external ear deformity and hypoplastic mandible. Case report. Scand. J. Plast. Reconstr. Surg. 1978;12:165–167. - PubMed
-
- Jampol M., Repetto G., Keith D.A., Curtin H., Remensynder J., Holmes L.B. New syndrome? Prominent, constricted ears with malformed condyle of the mandible. Am. J. Med. Genet. 1998;75:449–452. - PubMed
-
- Erlich M.S., Cunningham M.L., Hudgins L. Transmission of the dysgnathia complex from mother to daughter. Am. J. Med. Genet. 2000;95:269–274. - PubMed
-
- Guion-Almeida M.L., Zechi-Ceide R.M., Vendramini S., Kokitsu-Nakata N.M. Auriculo-condylar syndrome: additional patients. Am. J. Med. Genet. 2002;112:209–214. - PubMed
-
- Storm A.L., Johnson J.M., Lammer E., Green G.E., Cunniff C. Auriculo-condylar syndrome is associated with highly variable ear and mandibular defects in multiple kindreds. Am. J. Med. Genet. A. 2005;138A:141–145. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- UC2 HL103010/HL/NHLBI NIH HHS/United States
- RC2 HG005608/HG/NHGRI NIH HHS/United States
- RC2 HL102926/HL/NHLBI NIH HHS/United States
- HL103010/HL/NHLBI NIH HHS/United States
- RC2 HL102924/HL/NHLBI NIH HHS/United States
- HL102926/HL/NHLBI NIH HHS/United States
- (HL102923/HL/NHLBI NIH HHS/United States
- RC2 HL103010/HL/NHLBI NIH HHS/United States
- RC2 HL102923/HL/NHLBI NIH HHS/United States
- UC2 HL102926/HL/NHLBI NIH HHS/United States
- HL102925/HL/NHLBI NIH HHS/United States
- P30-DC05188/DC/NIDCD NIH HHS/United States
- HL102924/HL/NHLBI NIH HHS/United States
- P30 DC005188/DC/NIDCD NIH HHS/United States
- UC2 HL102923/HL/NHLBI NIH HHS/United States
- HG005608/HG/NHGRI NIH HHS/United States
- UC2 HL102924/HL/NHLBI NIH HHS/United States
- RC2 HL102925/HL/NHLBI NIH HHS/United States
- UC2 HL102925/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

