Catalytic mechanisms for phosphotriesterases
- PMID: 22561533
- PMCID: PMC3421070
- DOI: 10.1016/j.bbapap.2012.04.004
Catalytic mechanisms for phosphotriesterases
Abstract
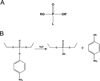
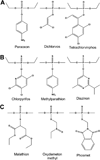
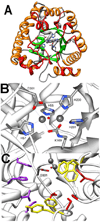
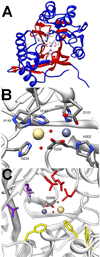
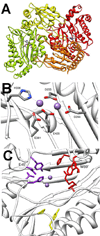
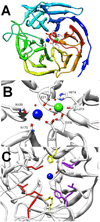
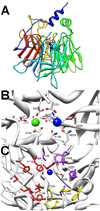
Phosphotriesters are one class of highly toxic synthetic compounds known as organophosphates. Wide spread usage of organophosphates as insecticides as well as nerve agents has lead to numerous efforts to identify enzymes capable of detoxifying them. A wide array of enzymes has been found to have phosphotriesterase activity including phosphotriesterase (PTE), methyl parathion hydrolase (MPH), organophosphorus acid anhydrolase (OPAA), diisopropylfluorophosphatase (DFP), and paraoxonase 1 (PON1). These enzymes differ widely in protein sequence and three-dimensional structure, as well as in catalytic mechanism, but they also share several common features. All of the enzymes identified as phosphotriesterases are metal-dependent hydrolases that contain a hydrophobic active site with three discrete binding pockets to accommodate the substrate ester groups. Activation of the substrate phosphorus center is achieved by a direct interaction between the phosphoryl oxygen and a divalent metal in the active site. The mechanistic details of the hydrolytic reaction differ among the various enzymes with both direct attack of a hydroxide as well as covalent catalysis being found. This article is part of a Special Issue entitled: Chemistry and mechanism of phosphatases, diesterases and triesterases.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures


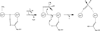

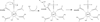
References
-
- Raushel FM. Bacterial detoxification of organophosphate nerve agents. Curr. Opin. Microbiol. 2002;5:288–295. - PubMed
-
- Musilek K, Dolezal M, Gunn-Moore F, Kuca K. Design, evaluation and structure---Activity relationship studies of the AChE reactivators against organophosphorus pesticides. Med. Res Rev. 2011;31:548–575. - PubMed
-
- Sun L, Dong Y, Zhou Y, Yang M, Zhang C, Rao Z, Zhang X. Crystalization and preliminary X-ray studies of methyl parathion hydrolase from Pseudomonas sp. WBC-3. Acta Crystallogr. D Biol. Crystallogr. 2004;60:954–956. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

