A cytochrome b5-containing plastid-located fatty acid desaturase from Chlamydomonas reinhardtii
- PMID: 22562471
- PMCID: PMC3416504
- DOI: 10.1128/EC.00079-12
A cytochrome b5-containing plastid-located fatty acid desaturase from Chlamydomonas reinhardtii
Abstract
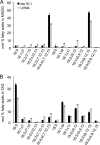
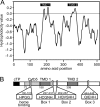
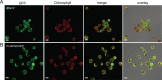
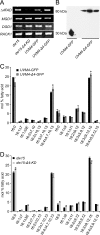
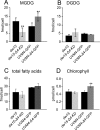
Monogalactosyldiacylglycerol (MGDG) in Chlamydomonas reinhardtii and other green algae contains hexadeca-4,7,10,13-tetraenoic acid (16:4) in the glycerol sn-2 position. While many genes necessary for the introduction of acyl chain double bonds have been functionally characterized, the Δ4-desaturase remained unknown. Using a phylogenetic comparison, a candidate gene encoding the MGDG-specific Δ4-desaturase from Chlamydomonas (CrΔ4FAD) was identified. CrΔ4FAD shows all characteristic features of a membrane-bound desaturase, including three histidine boxes and a transit peptide for chloroplast targeting. But it also has an N-terminal cytochrome b(5) domain, distinguishing it from other known plastid desaturases. Cytochrome b(5) is the primary electron donor for endoplasmic reticulum (ER) desaturases and is often fused to the desaturase domain in desaturases modifying the carboxyl end of the acyl group. Difference absorbance spectra of the recombinant cytochrome b(5) domain of CrΔ4FAD showed that it is functional in vitro. Green fluorescent protein fusions of CrΔ4FAD localized to the plastid envelope in Chlamydomonas. Interestingly, overproduction of CrΔ4FAD in Chlamydomonas not only increased levels of 16:4 acyl groups in cell extracts but specifically increased the total amount of MGDG. Vice versa, the amount of MGDG was lowered in lines with reduced levels of CrΔ4FAD. These data suggest a link between MGDG molecular species composition and galactolipid abundance in the alga, as well as a specific function for this fatty acid in MGDG.
Figures

References
-
- Abramoff M, Magelhaes P, Ram S. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 - PubMed
-
- Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

