Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle
- PMID: 22562991
- PMCID: PMC3416373
- DOI: 10.1128/AEM.07759-11
Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle
Abstract
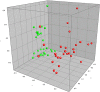
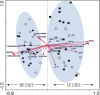
Feed-efficient animals have lower production costs and reduced environmental impact. Given that rumen microbial fermentation plays a pivotal role in host nutrition, the premise that rumen microbiota may contribute to host feed efficiency is gaining momentum. Since diet is a major factor in determining rumen community structure and fermentation patterns, we investigated the effect of divergence in phenotypic residual feed intake (RFI) on ruminal community structure of beef cattle across two contrasting diets. PCR-denaturing gradient gel electrophoresis (DGGE) and quantitative PCR (qPCR) were performed to profile the rumen bacterial population and to quantify the ruminal populations of Entodinium spp., protozoa, Fibrobacter succinogenes, Ruminococcus flavefaciens, Ruminococcus albus, Prevotella brevis, the genus Prevotella, and fungi in 14 low (efficient)- and 14 high (inefficient)-RFI animals offered a low-energy, high-forage diet, followed by a high-energy, low-forage diet. Canonical correspondence and Spearman correlation analyses were used to investigate associations between physiological variables and rumen microbial structure and specific microbial populations, respectively. The effect of RFI on bacterial profiles was influenced by diet, with the association between RFI group and PCR-DGGE profiles stronger for the higher forage diet. qPCR showed that Prevotella abundance was higher (P < 0.0001) in inefficient animals. A higher (P < 0.0001) abundance of Entodinium and Prevotella spp. and a lower (P < 0.0001) abundance of Fibrobacter succinogenes were observed when animals were offered the low-forage diet. Thus, differences in the ruminal microflora may contribute to host feed efficiency, although this effect may also be modulated by the diet offered.
Figures
References
-
- Briesacher SL, et al. 1992. Use of DNA probes to monitor nutritional effects on ruminal prokaryotes and Fibrobacter succinogenes S85. J. Anim. Sci. 70: 289–295 - PubMed
-
- Bustin SA, et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55: 611–622 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

