In vivo information-guided prediction approach for assessing the risks of drug-drug interactions associated with circulating inhibitory metabolites
- PMID: 22563046
- PMCID: PMC3400792
- DOI: 10.1124/dmd.112.045799
In vivo information-guided prediction approach for assessing the risks of drug-drug interactions associated with circulating inhibitory metabolites
Abstract
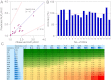
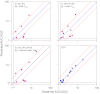
The in vivo drug-drug interaction (DDI) risks associated with cytochrome P450 inhibitors that have circulating inhibitory metabolites cannot be accurately predicted by conventional in vitro-based methods. A novel approach, in vivo information-guided prediction (IVIP), was recently introduced for CYP3A- and CYP2D6-mediated DDIs. This technique should be applicable to the prediction of DDIs involving other important cytochrome P450 metabolic pathways. Therefore, the aims of this study were to extend the IVIP approach to CYP2C9-mediated DDIs and evaluate the IVIP approach for predicting DDIs associated with inhibitory metabolites. The analysis was based on data from reported DDIs in the literature. The IVIP approach was modified and extended to CYP2C9-mediated DDIs. Thereafter, the IVIP approach was evaluated for predicting the DDI risks of various inhibitors with inhibitory metabolites. Although the data on CYP2C9-mediated DDIs were limited compared with those for CYP3A- and CYP2D6-mediated DDIs, the modified IVIP approach successfully predicted CYP2C9-mediated DDIs. For the external validation set, the prediction accuracy for area under the plasma concentration-time curve (AUC) ratios ranged from 70 to 125%. The accuracy (75-128%) of the IVIP approach in predicting DDI risks of inhibitors with circulating inhibitory metabolites was more accurate than in vitro-based methods (28-805%). The IVIP model accommodates important confounding factors in the prediction of DDIs, which are difficult to handle using in vitro-based methods. In conclusion, the IVIP approach could be used to predict CYP2C9-mediated DDIs and is easily modified to incorporate the additive effect of circulating inhibitory metabolites.
Figures
References
-
- Albers LJ, Reist C, Vu RL, Fujimoto K, Ozdemir V, Helmeste D, Poland R, Tang SW. (2000) Effect of venlafaxine on imipramine metabolism. Psychiatry Res 96:235–243 - PubMed
-
- Alderman J, Preskorn SH, Greenblatt DJ, Harrison W, Penenberg D, Allison J, Chung M. (1997) Desipramine pharmacokinetics when coadministered with paroxetine or sertraline in extensive metabolizers. J Clin Psychopharmacol 17:284–291 - PubMed
-
- Ayesh R, Dawling S, Hayler A, Oates NS, Cholerton S, Widdop B, Idle JR, Smith RL. (1991) Comparative effects of the diastereoisomers, quinine and quinidine in producing phenocopy debrisoquine poor metabolisers (PMs) in healthy volunteers. Chirality 3:14–18 - PubMed
-
- Backman JT, Kivistö KT, Olkkola KT, Neuvonen PJ. (1998) The area under the plasma concentration-time curve for oral midazolam is 400-fold larger during treatment with itraconazole than with rifampicin. Eur J Clin Pharmacol 54:53–58 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

