SalB inactivation modulates culture supernatant exoproteins and affects autolysis and viability in Enterococcus faecalis OG1RF
- PMID: 22563054
- PMCID: PMC3393493
- DOI: 10.1128/JB.00376-12
SalB inactivation modulates culture supernatant exoproteins and affects autolysis and viability in Enterococcus faecalis OG1RF
Abstract
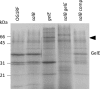
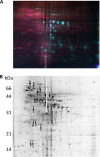
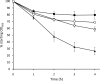
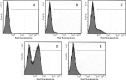
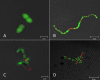
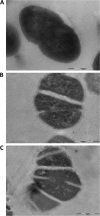
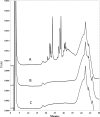
The culture supernatant fraction of an Enterococcus faecalis gelE mutant of strain OG1RF contained elevated levels of the secreted antigen SalB. Using differential fluorescence gel electrophoresis (DIGE) the salB mutant was shown to possess a unique complement of exoproteins. Differentially abundant exoproteins were identified using matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry. Stress-related proteins including DnaK, Dps family protein, SOD, and NADH peroxidase were present in greater quantity in the OG1RF salB mutant culture supernatant. Moreover, several proteins involved in cell wall synthesis and cell division, including d-Ala-d-Lac ligase and EzrA, were present in reduced quantity in OG1RF salB relative to the parent strain. The salB mutant displayed reduced viability and anomalous cell division, and these phenotypes were exacerbated in a gelE salB double mutant. An epistatic relationship between gelE and salB was not identified with respect to increased autolysis and cell morphological changes observed in the salB mutant. SalB was purified as a six-histidine-tagged protein to investigate peptidoglycan hydrolytic activity; however, activity was not evident. High-pressure liquid chromatography (HPLC) analysis of reduced muropeptides from peptidoglycan digested with mutanolysin revealed that the salB mutant and OG1RF were indistinguishable.
Figures
References
-
- Arias CA, Cortes L, Murray BE. 2007. Chaining in enterococci revisited: correlation between chain length and gelatinase phenotype, and gelE and fsrB genes among clinical isolates of Enterococcus faecalis. J. Med. Microbiol. 56:286–288 - PubMed
-
- Biavasco F, et al. 1996. In vitro conjugative transfer of VanA vancomycin resistance between enterococci and listeriae of different species. Eur. J. Clin. Microbiol. Infect. Dis. 15:50–59 - PubMed
-
- Bonten MJ, Willems R, Weinstein RA. 2001. Vancomycin-resistant enterococci: why are they here and where do they come from? Lancet Infect. Dis. 1:314–325 - PubMed
-
- Breton YL, et al. 2002. Isolation and characterization of bile salts-sensitive mutants of Enterococcus faecalis. Curr. Microbiol. 45:434–439 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

