Beyond modeling: all-atom olfactory receptor model simulations
- PMID: 22563330
- PMCID: PMC3342527
- DOI: 10.3389/fgene.2012.00061
Beyond modeling: all-atom olfactory receptor model simulations
Abstract
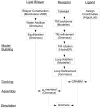
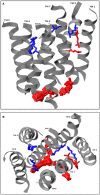
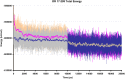
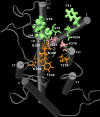
Olfactory receptors (ORs) are a type of GTP-binding protein-coupled receptor (GPCR). These receptors are responsible for mediating the sense of smell through their interaction with odor ligands. OR-odorant interactions marks the first step in the process that leads to olfaction. Computational studies on model OR structures can generate focused and novel hypotheses for further bench investigation by providing a view of these interactions at the molecular level beyond inferences that are drawn merely from static docking. Here we have shown the specific advantages of simulating the dynamic environment associated with OR-odorant interactions. We present a rigorous protocol which ranges from the creation of a computationally derived model of an olfactory receptor to simulating the interactions between an OR and an odorant molecule. Given the ubiquitous occurrence of GPCRs in the membranes of cells, we anticipate that our OR-developed methodology will serve as a model for the computational structural biology of all GPCRs.
Keywords: GPCR; ligand binding; lipid bilayer; molecular dynamics; olfactory receptor; protein modeling.
Figures


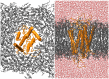
References
Grants and funding
LinkOut - more resources
Full Text Sources

