The neutrophil's eye-view: inference and visualisation of the chemoattractant field driving cell chemotaxis in vivo
- PMID: 22563379
- PMCID: PMC3338515
- DOI: 10.1371/journal.pone.0035182
The neutrophil's eye-view: inference and visualisation of the chemoattractant field driving cell chemotaxis in vivo
Abstract
As we begin to understand the signals that drive chemotaxis in vivo, it is becoming clear that there is a complex interplay of chemotactic factors, which changes over time as the inflammatory response evolves. New animal models such as transgenic lines of zebrafish, which are near transparent and where the neutrophils express a green fluorescent protein, have the potential to greatly increase our understanding of the chemotactic process under conditions of wounding and infection from video microscopy data. Measurement of the chemoattractants over space (and their evolution over time) is a key objective for understanding the signals driving neutrophil chemotaxis. However, it is not possible to measure and visualise the most important contributors to in vivo chemotaxis, and in fact the understanding of the main contributors at any particular time is incomplete. The key insight that we make in this investigation is that the neutrophils themselves are sensing the underlying field that is driving their action and we can use the observations of neutrophil movement to infer the hidden net chemoattractant field by use of a novel computational framework. We apply the methodology to multiple in vivo neutrophil recruitment data sets to demonstrate this new technique and find that the method provides consistent estimates of the chemoattractant field across the majority of experiments. The framework that we derive represents an important new methodology for cell biologists investigating the signalling processes driving cell chemotaxis, which we label the neutrophils eye-view of the chemoattractant field.
Conflict of interest statement
Figures


 1000 pixels has been zoomed on the vertical axis to the 100–900 pixel range.
1000 pixels has been zoomed on the vertical axis to the 100–900 pixel range.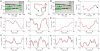

 m/min range for an effective visualisation and data are aggregated over all fish). B: Histogram of neutrophil velocities in the Y-direction.
m/min range for an effective visualisation and data are aggregated over all fish). B: Histogram of neutrophil velocities in the Y-direction.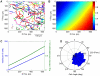

References
-
- Porter SL, Wadhams GH, Armitage JP. Signal processing in complex chemotaxis pathways. Nature Reviews Microbiology. 2011;9:153–165. - PubMed
-
- Parent CA. Making all the right moves: chemotaxis in neutrophils and dictyostelium. Current Opinion in Cell Biology. 2004;16:4–13. - PubMed
-
- Nathan C. Neutrophils and immunity: challenges and opportunities. Nature Reviews Immunology. 2006;6:173–182. - PubMed
-
- Silva MT. Macrophage phagocytosis of neutrophils at inammatory/infectious foci: a cooperative mechanism in the control of infection and infectious inammation. Journal of Leukocyte Biology. 2011;89:675–683. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

