Clusters of ancestrally related genes that show paralogy in whole or in part are a major feature of the genomes of humans and other species
- PMID: 22563380
- PMCID: PMC3338513
- DOI: 10.1371/journal.pone.0035274
Clusters of ancestrally related genes that show paralogy in whole or in part are a major feature of the genomes of humans and other species
Abstract
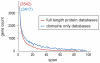
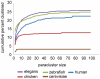
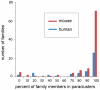
Arrangements of genes along chromosomes are a product of evolutionary processes, and we can expect that preferable arrangements will prevail over the span of evolutionary time, often being reflected in the non-random clustering of structurally and/or functionally related genes. Such non-random arrangements can arise by two distinct evolutionary processes: duplications of DNA sequences that give rise to clusters of genes sharing both sequence similarity and common sequence features and the migration together of genes related by function, but not by common descent. To provide a background for distinguishing between the two, which is important for future efforts to unravel the evolutionary processes involved, we here provide a description of the extent to which ancestrally related genes are found in proximity.Towards this purpose, we combined information from five genomic datasets, InterPro, SCOP, PANTHER, Ensembl protein families, and Ensembl gene paralogs. The results are provided in publicly available datasets (http://cgd.jax.org/datasets/clustering/paraclustering.shtml) describing the extent to which ancestrally related genes are in proximity beyond what is expected by chance (i.e. form paraclusters) in the human and nine other vertebrate genomes, as well as the D. melanogaster, C. elegans, A. thaliana, and S. cerevisiae genomes. With the exception of Saccharomyces, paraclusters are a common feature of the genomes we examined. In the human genome they are estimated to include at least 22% of all protein coding genes. Paraclusters are far more prevalent among some gene families than others, are highly species or clade specific and can evolve rapidly, sometimes in response to environmental cues. Altogether, they account for a large portion of the functional clustering previously reported in several genomes.
Conflict of interest statement
Figures



References
-
- Fisher RA. 1930. The Genetic Theory of Natural Selection, Clarendon Press, Oxford, UK.
-
- Nei M. Genome evolution: let's stick together. Heredity. 2003;90:411–412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

