Environment Polarity in Proteins Mapped Noninvasively by FTIR Spectroscopy
- PMID: 22563521
- PMCID: PMC3341589
- DOI: 10.1021/jz300150v
Environment Polarity in Proteins Mapped Noninvasively by FTIR Spectroscopy
Abstract
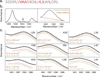
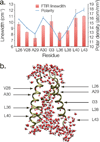
The polarity pattern of a macromolecule is of utmost importance to its structure and function. For example, one of the main driving forces for protein folding is the burial of hydrophobic residues. Yet polarity remains a difficult property to measure experimentally, due in part to its non-uniformity in the protein interior. Herein, we show that FTIR linewidth analysis of noninvasive 1-(13)C=(18)O labels can be used to obtain a reliable measure of the local polarity, even in a highly multi-phasic system, such as a membrane protein. We show that in the Influenza M2 H(+) channel, residues that line the pore are located in an environment that is as polar as fully solvated residues, while residues that face the lipid acyl chains are located in an apolar environment. Taken together, FTIR linewidth analysis is a powerful, yet chemically non-perturbing approach to examine one of the most important properties in proteins - polarity.
Figures
References
-
- Langmuir I. Protein Monolayers. Cold Spring Harbor Symp Quant Biol. 1938;6:171–189.
-
- Pey AL, Rodriguez-Larrea D, Gavira JA, Garcia-Moreno B, Sanchez-Ruiz JM. Modulation of Buried Ionizable Groups In Proteins With Engineered Surface Charge. J Am Chem Soc. 2010;132:1218–1219. - PubMed
-
- Getahun Z, Huang C-Y, Wang T, De León B, DeGrado WF, Gai F. Using Nitrile-derivatized Amino Acids as Infrared Probes of Local Environment. J Am Chem Soc. 2003;125:405–411. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

