Microarray-based uncovering reference genes for quantitative real time PCR in grapevine under abiotic stress
- PMID: 22564373
- PMCID: PMC3837474
- DOI: 10.1186/1756-0500-5-220
Microarray-based uncovering reference genes for quantitative real time PCR in grapevine under abiotic stress
Abstract
Background: Quantitative real time polymerase chain reaction is becoming the primary tool for detecting mRNA and transcription data analysis as it shows to have advantages over other more commonly used techniques. Nevertheless, it also presents a few shortcomings, with the most import being the need for data normalisation, usually with a reference gene. Therefore the choice of the reference gene(s) is of great importance for correct data analysis. Microarray data, when available, can be of great assistance when choosing reference genes. Grapevine was submitted to water stress and heat stress as well as a combination of both to test the stability of the possible reference genes.
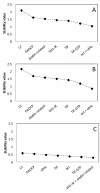
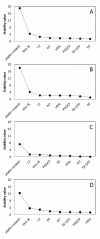
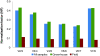
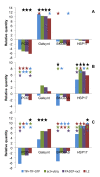
Results: Using the analysis of microarray data available for grapevine, six possible reference genes were selected for RT-qPCR validation: PADCP, ubiq, TIF, TIF-GTP, VH1-IK, aladin-related. Two additional genes that are commonly used as reference genes were included: act and L2. The stability of those genes was tested in leaves of grapevine in both field plants and in greenhouse plants under water or heat stress or a combination of both. Gene stability was analyzed with the softwares GeNorm, NormFinder and the ΔCq method resulting in several combinations of reference genes suitable for data normalisation. In order to assess the best combination, the reference genes were tested in putative stress marker genes (PCO, Galsynt, BKCoAS and HSP17) also chosen from the same microarray, in water stress, heat stress and the combination of both.
Conclusions: Each method selected different gene combinations (PADCP + act, TIF + TIF-GTP and ubiq + act). However, as none of the combinations diverged significantly from the others used to normalize the expression of the putative stress marker genes, then any combination is suitable for data normalisation under the conditions tested. Here we prove the accuracy of choosing grapevine reference genes for RT-qPCR through a microarray analysis.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

