CDK targeting of NBS1 promotes DNA-end resection, replication restart and homologous recombination
- PMID: 22565321
- PMCID: PMC3367243
- DOI: 10.1038/embor.2012.58
CDK targeting of NBS1 promotes DNA-end resection, replication restart and homologous recombination
Abstract
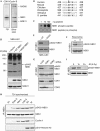
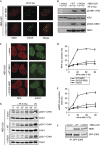
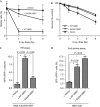
The conserved MRE11–RAD50–NBS1 (MRN) complex is an important sensor of DNA double-strand breaks (DSBs) and facilitates DNA repair by homologous recombination (HR) and end joining. Here, we identify NBS1 as a target of cyclin-dependent kinase (CDK) phosphorylation. We show that NBS1 serine 432 phosphorylation occurs in the S, G2 and M phases of the cell cycle and requires CDK activity. This modification stimulates MRN-dependent conversion of DSBs into structures that are substrates for repair by HR. Impairment of NBS1 phosphorylation not only negatively affects DSB repair by HR, but also prevents resumption of DNA replication after replication-fork stalling. Thus, CDK-mediated NBS1 phosphorylation defines a molecular switch that controls the choice of repair mode for DSBs.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28: 739–745 - PubMed
-
- Falck J, Coates J, Jackson SP (2005) Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434: 605–611 - PubMed
-
- Matsuoka S et al. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

